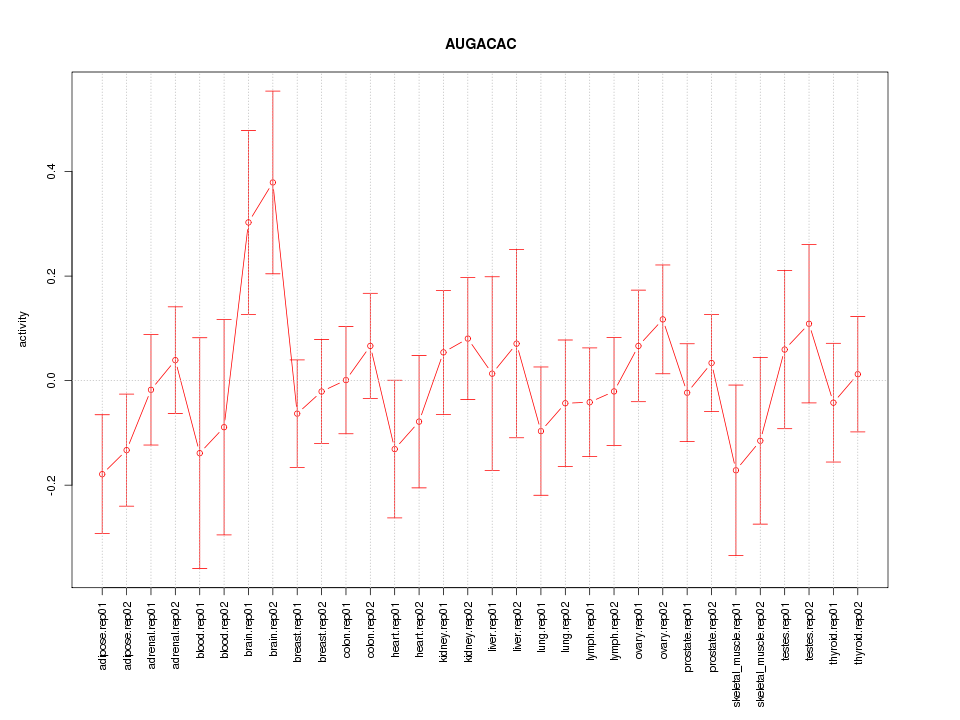
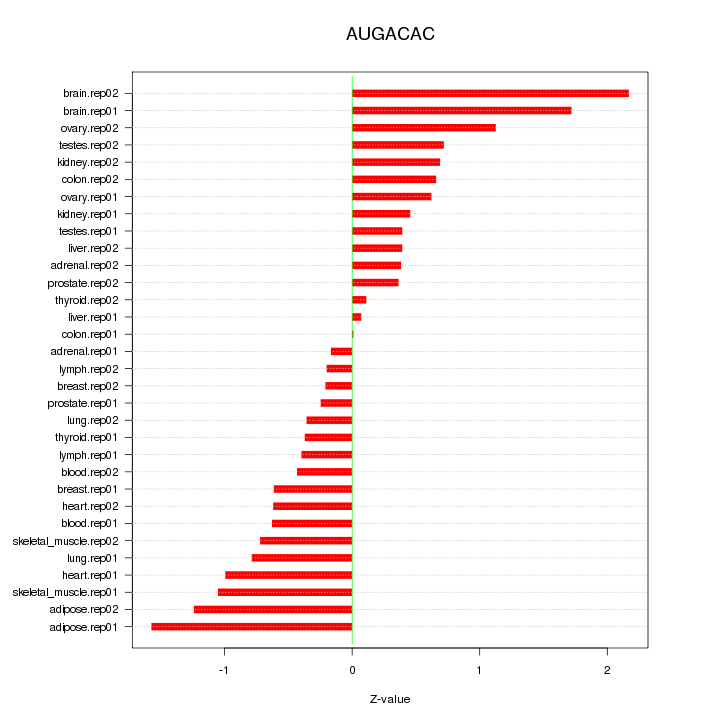
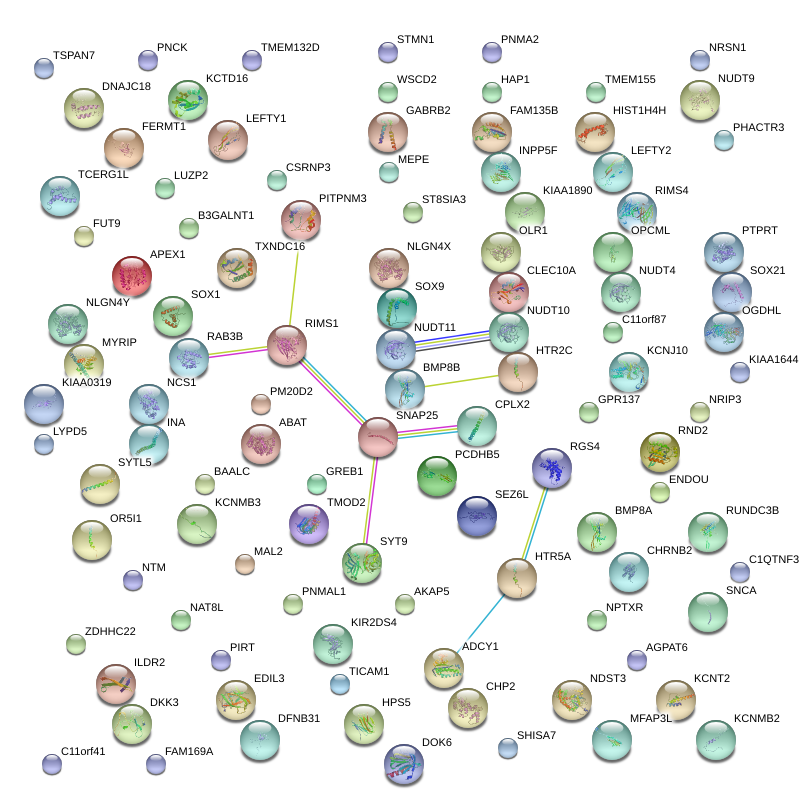
|
chr19_-_55954229
|
2.951
|
NM_001145176
|
SHISA7
|
shisa homolog 7 (Xenopus laevis)
|
|
chr5_-_74162614
|
1.916
|
NM_015566
|
FAM169A
|
family with sequence similarity 169, member A
|
|
chr1_-_52456292
|
1.874
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr22_-_44708730
|
1.665
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr7_+_45614081
|
1.622
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr20_+_10199455
|
1.553
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr11_+_33563772
|
1.534
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr9_+_132934685
|
1.478
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr12_-_130388211
|
1.465
|
NM_133448
|
TMEM132D
|
transmembrane protein 132D
|
|
chr1_-_160039946
|
1.384
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr10_+_105036783
|
1.364
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr9_+_132962871
|
1.338
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr1_+_163038934
|
1.312
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr18_+_55019720
|
1.214
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr6_+_24126349
|
1.132
|
NM_080723
|
NRSN1
|
neurensin 1
|
|
chr4_+_2061238
|
1.120
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr5_+_175223519
|
1.113
|
NM_006650
|
CPLX2
|
complexin 2
|
|
chr8_-_26371387
|
1.113
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr1_+_163038564
|
1.091
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chrX_+_51075082
|
1.076
|
NM_153183
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr13_+_37248048
|
1.008
|
NM_203451
|
SERTM1
|
serine-rich and transmembrane domain containing 1
|
|
chr14_-_77608132
|
1.006
|
NM_174976
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr7_+_154862545
|
0.982
|
NM_024012
|
HTR5A
|
5-hydroxytryptamine (serotonin) receptor 5A
|
|
chr17_-_6459746
|
0.967
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr5_+_175298492
|
0.962
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr17_+_41177247
|
0.948
|
NM_005440
|
RND2
|
Rho family GTPase 2
|
|
chr8_+_120220554
|
0.913
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr4_-_122686281
|
0.909
|
NM_152399
|
TMEM155
|
transmembrane protein 155
|
|
chr1_+_163041694
|
0.900
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr10_-_50970321
|
0.895
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr22_+_26565424
|
0.892
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr1_+_163038395
|
0.873
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr18_+_67068134
|
0.863
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr5_+_140514787
|
0.843
|
NM_015669
|
PCDHB5
|
protocadherin beta 5
|
|
chr11_-_9025531
|
0.842
|
NM_020645
|
NRIP3
|
nuclear receptor interacting protein 3
|
|
chr11_+_109292837
|
0.809
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr1_-_166944504
|
0.809
|
NM_199351
|
ILDR2
|
immunoglobulin-like domain containing receptor 2
|
|
chr9_-_117267710
|
0.793
|
NM_001173425
NM_015404
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr17_-_10741398
|
0.793
|
NM_001101387
|
PIRT
|
phosphoinositide-interacting regulator of transient receptor potential channels
|
|
chr11_+_7273141
|
0.790
|
NM_175733
|
SYT9
|
synaptotagmin IX
|
|
chr8_-_139509064
|
0.776
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr6_-_24645955
|
0.775
|
NM_001168375
NM_001168376
NM_001168374
NM_001168377
NM_014809
|
KIAA0319
|
KIAA0319
|
|
chr2_+_166326156
|
0.769
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr5_+_143550395
|
0.768
|
NM_020768
|
KCTD16
|
potassium channel tetramerisation domain containing 16
|
|
chr11_-_133402227
|
0.753
|
NM_001012393
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr19_-_46974789
|
0.751
|
NM_001103149
NM_018215
|
PNMAL1
|
PNMA-like 1
|
|
chr1_-_26232643
|
0.750
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr5_-_160975124
|
0.749
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr5_-_138775213
|
0.744
|
NM_152686
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chr2_+_166428845
|
0.743
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr20_+_58251715
|
0.737
|
NM_001199506
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr17_-_39890882
|
0.736
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr14_+_64932216
|
0.728
|
NM_004857
|
AKAP5
|
A kinase (PRKA) anchor protein 5
|
|
chr13_-_95364248
|
0.726
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr16_+_8768403
|
0.723
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr11_-_132812870
|
0.706
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr6_+_96463844
|
0.702
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chrX_-_6146655
|
0.692
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr1_+_154540250
|
0.691
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr20_-_31239558
|
0.687
|
NM_182584
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr20_+_58179601
|
0.684
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr3_+_159943252
|
0.683
|
NM_001168214
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr20_+_58296264
|
0.671
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr20_+_58152563
|
0.662
|
NM_001199505
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr12_+_108523464
|
0.654
|
NM_014653
|
WSCD2
|
WSC domain containing 2
|
|
chr15_+_52043738
|
0.653
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr20_-_41818464
|
0.649
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr12_-_10324675
|
0.648
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr4_+_88754120
|
0.648
|
NM_001184695
NM_001184696
NM_001184697
NM_020203
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chrX_-_152939764
|
0.636
|
NM_001039582
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr3_+_39851028
|
0.633
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chrX_+_113818550
|
0.633
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chrX_+_37865834
|
0.632
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr17_+_76227390
|
0.626
|
NM_001204210
NM_001204211
NM_001204212
|
TMEM235
|
transmembrane protein 235
|
|
chr4_-_90759446
|
0.625
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr4_+_118955499
|
0.622
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr20_-_43438881
|
0.619
|
NM_001205317
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr2_+_11674241
|
0.607
|
NM_014668
|
GREB1
|
growth regulation by estrogen in breast cancer 1
|
|
chrX_-_6145887
|
0.594
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr10_-_133109983
|
0.594
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr11_+_24518515
|
0.585
|
NM_001009909
NM_001252008
NM_001252010
|
LUZP2
|
leucine zipper protein 2
|
|
chr4_-_170947428
|
0.585
|
NM_021647
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr19_-_44324767
|
0.579
|
NM_182573
|
LYPD5
|
LY6/PLAUR domain containing 5
|
|
chr16_+_23765947
|
0.577
|
NM_022097
|
CHP2
|
calcineurin B homologous protein 2
|
|
chr7_+_87257704
|
0.576
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr1_-_226076766
|
0.574
|
NM_020997
|
LEFTY1
|
left-right determination factor 1
|
|
chr6_+_72922513
|
0.573
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr11_-_12030882
|
0.570
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_-_26233367
|
0.568
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_+_39957298
|
0.555
|
NM_181809
|
BMP8A
|
bone morphogenetic protein 8a
|
|
chr17_+_70117154
|
0.554
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr11_-_12030622
|
0.553
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr3_-_160822625
|
0.550
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr9_-_117265452
|
0.545
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr1_-_196577438
|
0.531
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr12_-_48119224
|
0.528
|
NM_001172439
NM_001172440
NM_006025
|
ENDOU
|
endonuclease, polyU-specific
|
|
chr5_-_83680590
|
0.527
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr5_-_34043234
|
0.527
|
NM_030945
NM_181435
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3
|
|
chr20_-_6104056
|
0.522
|
NM_017671
|
FERMT1
|
fermitin family member 1
|
|
chr22_-_39240016
|
0.518
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chrX_+_38420690
|
0.517
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr8_+_104152876
|
0.515
|
NM_001024372
NM_024812
|
BAALC
|
brain and acute leukemia, cytoplasmic
|
|
chr3_+_178276487
|
0.515
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr16_+_8814567
|
0.514
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr14_-_88737160
|
0.506
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_-_12030128
|
0.503
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr1_+_226736500
|
0.501
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr4_-_156298094
|
0.500
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr20_-_36889173
|
0.493
|
NM_001029864
|
KIAA1755
|
KIAA1755
|
|
chr22_+_35937351
|
0.488
|
NM_014310
|
RASD2
|
RASD family, member 2
|
|
chr7_+_79764087
|
0.486
|
NM_002069
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1
|
|
chr11_-_6502563
|
0.486
|
NM_001242854
NM_001242855
NM_001242856
NM_012402
|
ARFIP2
|
ADP-ribosylation factor interacting protein 2
|
|
chr4_-_17513676
|
0.480
|
NM_000320
|
QDPR
|
quinoid dihydropteridine reductase
|
|
chr11_-_74109417
|
0.470
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr8_+_91803777
|
0.469
|
NM_022351
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1
|
|
chr7_+_128828712
|
0.466
|
NM_005631
|
SMO
|
smoothened, frizzled family receptor
|
|
chr19_-_44306525
|
0.464
|
NM_001031749
|
LYPD5
|
LY6/PLAUR domain containing 5
|
|
chr4_-_90758126
|
0.461
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr14_-_94254765
|
0.455
|
NM_178013
|
PRIMA1
|
proline rich membrane anchor 1
|
|
chr12_+_1929432
|
0.455
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr1_-_184006828
|
0.452
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr14_-_88789531
|
0.451
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr12_-_104234968
|
0.448
|
NM_001031701
|
NT5DC3
|
5'-nucleotidase domain containing 3
|
|
chr11_-_123525300
|
0.447
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr8_-_33330630
|
0.446
|
NM_032664
|
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase)
|
|
chr20_+_8112762
|
0.444
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr15_+_69606741
|
0.441
|
NM_001104554
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr2_-_73496595
|
0.433
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr12_+_72666462
|
0.427
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr15_+_69591272
|
0.421
|
NM_017705
|
PAQR5
|
progestin and adipoQ receptor family member V
|
|
chr16_+_18995255
|
0.421
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr1_-_241803203
|
0.419
|
NM_014322
|
OPN3
|
opsin 3
|
|
chr16_+_8806825
|
0.419
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr4_-_96470350
|
0.416
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr4_+_88742549
|
0.413
|
NM_001184694
|
MEPE
|
matrix extracellular phosphoglycoprotein
|
|
chr18_-_47721165
|
0.409
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr12_+_57828457
|
0.405
|
NM_005538
|
INHBC
|
inhibin, beta C
|
|
chr4_-_170924911
|
0.403
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chrX_-_152938742
|
0.403
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr18_+_52495618
|
0.402
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr18_+_55102777
|
0.397
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr13_-_46425756
|
0.394
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr3_+_178254223
|
0.393
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr14_-_88793250
|
0.392
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_-_26593667
|
0.387
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr16_+_85061303
|
0.386
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr5_+_68800003
|
0.383
|
NM_001205255
|
OCLN
|
occludin
|
|
chr1_-_22469414
|
0.377
|
NM_030761
|
WNT4
|
wingless-type MMTV integration site family, member 4
|
|
chr8_+_75736771
|
0.377
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr6_+_73076431
|
0.373
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_-_7044664
|
0.373
|
NM_153376
|
CCDC96
|
coiled-coil domain containing 96
|
|
chrX_-_10851808
|
0.369
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr8_-_143695812
|
0.367
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr5_+_178368193
|
0.364
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr13_+_46039024
|
0.363
|
NM_031431
|
COG3
|
component of oligomeric golgi complex 3
|
|
chr17_-_27332888
|
0.362
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr6_+_72926462
|
0.360
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr5_+_156696351
|
0.359
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr14_-_93651242
|
0.359
|
NM_022151
|
MOAP1
|
modulator of apoptosis 1
|
|
chr10_-_129691202
|
0.359
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr5_+_156693051
|
0.359
|
NM_001037333
NM_001037332
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr15_+_51633712
|
0.359
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr16_+_18995508
|
0.356
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr1_+_154966061
|
0.356
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr1_+_28099682
|
0.355
|
NM_177424
|
STX12
|
syntaxin 12
|
|
chr15_-_52483564
|
0.354
|
NM_016194
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr19_+_11976843
|
0.354
|
NM_152262
|
ZNF439
|
zinc finger protein 439
|
|
chr3_-_178789597
|
0.353
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr18_-_29264394
|
0.353
|
NM_004775
|
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6
|
|
chr8_+_63161499
|
0.348
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr17_-_61523544
|
0.347
|
NM_001017916
NM_001915
|
CYB561
|
cytochrome b-561
|
|
chr11_+_121322848
|
0.347
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr1_+_2518071
|
0.345
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr4_+_37585854
|
0.342
|
NM_018302
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr6_-_41006927
|
0.340
|
NM_173561
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like
|
|
chr20_-_4990913
|
0.339
|
NM_203327
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr13_+_51796363
|
0.338
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr4_-_53525316
|
0.336
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr1_+_25943958
|
0.336
|
NM_020379
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1
|
|
chr9_+_133884503
|
0.335
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr17_+_53342320
|
0.333
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr6_+_158402887
|
0.333
|
NM_003898
|
SYNJ2
|
synaptojanin 2
|
|
chr13_+_24734788
|
0.332
|
NM_001166271
NM_153023
|
SPATA13
|
spermatogenesis associated 13
|
|
chr6_+_72926144
|
0.332
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr17_+_43861628
|
0.332
|
NM_001145146
NM_001145147
NM_001145148
NM_004382
|
CRHR1
|
corticotropin releasing hormone receptor 1
|
|
chr15_-_52472075
|
0.328
|
NM_006578
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr13_-_37494370
|
0.326
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr4_+_37455551
|
0.321
|
NM_001104629
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr6_+_157802479
|
0.320
|
NM_024630
NM_153746
|
ZDHHC14
|
zinc finger, DHHC-type containing 14
|
|
chr20_-_4982133
|
0.319
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr11_-_8832881
|
0.319
|
NM_139157
|
ST5
|
suppression of tumorigenicity 5
|
|
chr2_+_54952148
|
0.317
|
NM_001039753
|
EML6
|
echinoderm microtubule associated protein like 6
|
|
chr7_-_138665853
|
0.317
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr5_-_148442606
|
0.317
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr15_+_32322690
|
0.311
|
NM_000746
NM_001190455
|
CHRNA7
|
cholinergic receptor, nicotinic, alpha 7
|
|
chr11_+_13690151
|
0.306
|
NM_032228
|
FAR1
|
fatty acyl CoA reductase 1
|
|
chr13_-_30169795
|
0.300
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr15_-_56035176
|
0.299
|
NM_173814
|
PRTG
|
protogenin
|
|
chr8_-_88886224
|
0.298
|
NM_152418
|
DCAF4L2
|
DDB1 and CUL4 associated factor 4-like 2
|
|
chr2_+_26568900
|
0.298
|
NM_033505
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|