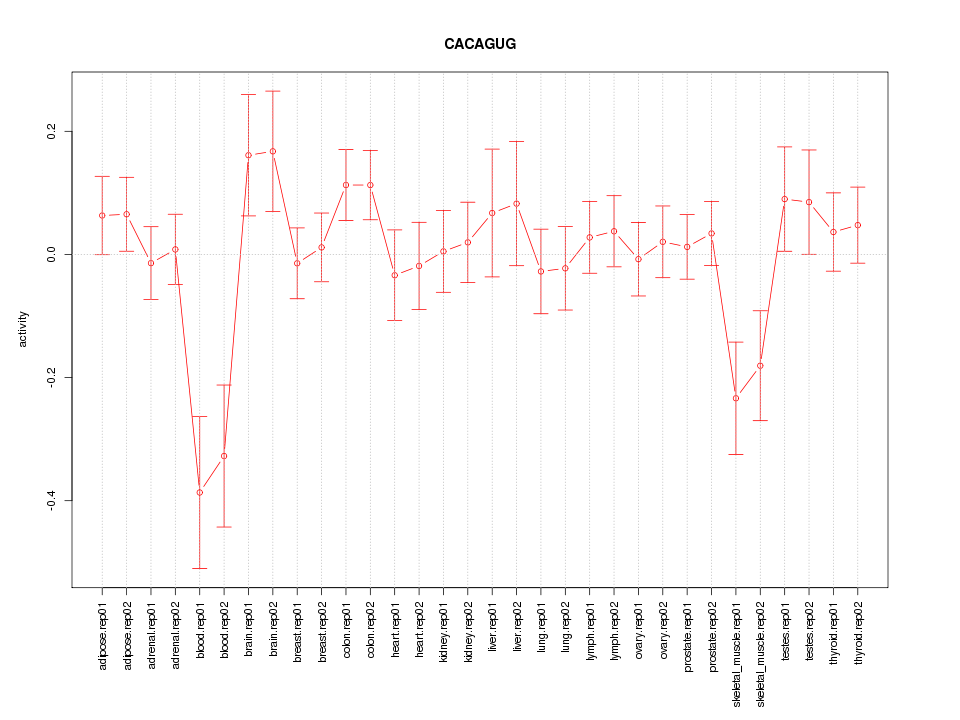
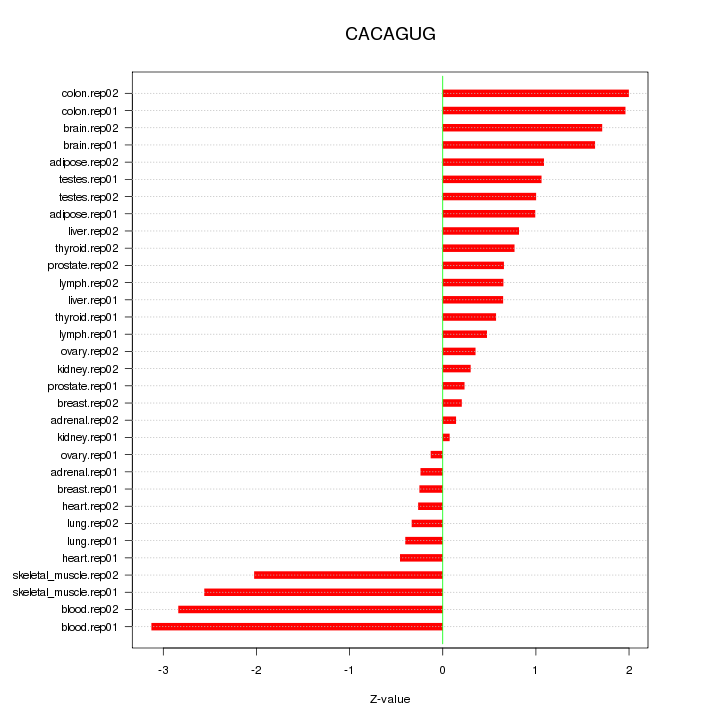
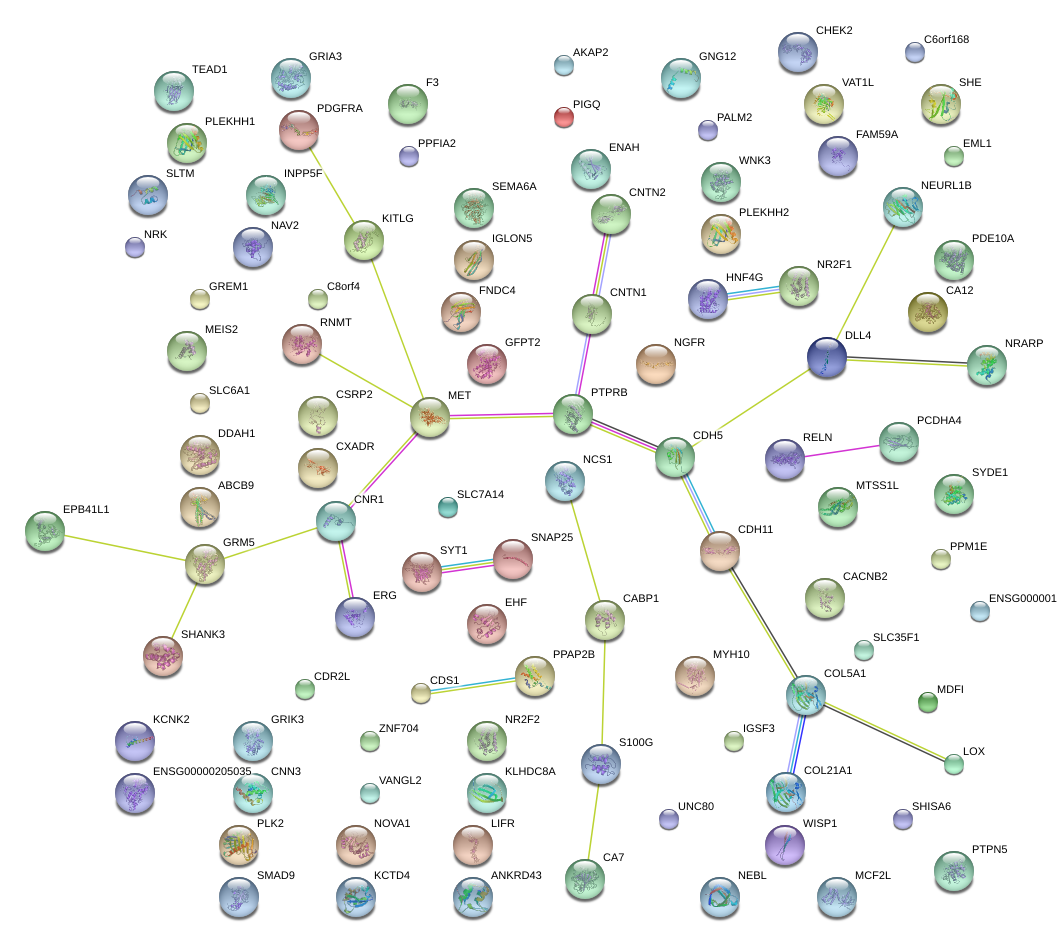
|
chr15_+_33010174
|
8.988
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr1_-_95007139
|
7.147
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr12_-_77272767
|
7.001
|
NM_001321
|
CSRP2
|
cysteine and glycine-rich protein 2
|
|
chr19_+_51815095
|
6.245
|
NM_001101372
|
IGLON5
|
IgLON family member 5
|
|
chr9_+_132934685
|
5.825
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr1_-_68299047
|
5.751
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr5_-_38595506
|
5.582
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr2_-_27718098
|
5.515
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr14_+_100259445
|
5.276
|
NM_001008707
NM_004434
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr5_-_38556682
|
5.208
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr22_+_51113069
|
5.091
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr1_-_86043932
|
5.072
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_85930733
|
5.014
|
NM_012137
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr20_+_10199455
|
4.996
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr9_+_132962871
|
4.981
|
NM_001128826
|
NCS1
|
neuronal calcium sensor 1
|
|
chr6_-_166075520
|
4.612
|
NM_006661
NM_001130690
|
PDE10A
|
phosphodiesterase 10A
|
|
chr6_+_41606193
|
4.532
|
NM_005586
|
MDFI
|
MyoD family inhibitor
|
|
chr12_+_121088370
|
4.497
|
NM_004276
NM_031205
|
CABP1
|
calcium binding protein 1
|
|
chr12_+_79439432
|
4.464
|
NM_001135806
|
SYT1
|
synaptotagmin I
|
|
chr11_+_34642587
|
4.393
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr15_-_37393364
|
4.346
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr5_+_132149022
|
4.306
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr7_-_28998028
|
4.303
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr13_-_37494370
|
4.299
|
NM_001127217
NM_005905
|
SMAD9
|
SMAD family member 9
|
|
chr8_+_40010986
|
4.266
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr7_-_103629962
|
4.180
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr5_-_57755787
|
4.142
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr1_+_160370363
|
4.120
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr12_+_79257772
|
4.119
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr5_-_115910406
|
4.082
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr16_-_70719854
|
4.079
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr11_+_12695851
|
4.053
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr4_+_85504056
|
4.039
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr7_+_116312412
|
4.034
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr1_-_154474524
|
4.029
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chrX_+_122318095
|
3.942
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr21_-_39870303
|
3.895
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_+_19734821
|
3.825
|
NM_001244963
NM_145117
NM_182964
|
NAV2
|
neuron navigator 2
|
|
chr16_+_66400524
|
3.822
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr17_-_8534008
|
3.789
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr15_-_37392358
|
3.783
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr12_+_79258448
|
3.770
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr10_+_18429605
|
3.755
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr6_-_88875766
|
3.751
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr3_-_170303774
|
3.733
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr15_+_96875793
|
3.648
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr16_+_77822400
|
3.516
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chrX_+_105066531
|
3.513
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr17_+_56833231
|
3.511
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr1_-_37499708
|
3.508
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr16_-_65155881
|
3.492
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr17_+_47572592
|
3.490
|
NM_002507
|
NGFR
|
nerve growth factor receptor
|
|
chr8_-_81787015
|
3.451
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr11_-_18813127
|
3.450
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr12_+_121078421
|
3.426
|
NM_001033677
|
CABP1
|
calcium binding protein 1
|
|
chr1_-_95392501
|
3.392
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr19_+_15218141
|
3.314
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr5_-_121412805
|
3.280
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr20_+_34700257
|
3.264
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr6_+_118228688
|
3.249
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr20_+_24449834
|
3.245
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr8_+_76452202
|
3.230
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr20_+_34742656
|
3.183
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr17_+_11144739
|
3.175
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr11_-_88781039
|
3.163
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr1_-_57045235
|
3.121
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr15_+_96873845
|
3.119
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_+_72983723
|
3.092
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr14_-_27066959
|
3.036
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr12_-_123451031
|
3.000
|
NM_001243013
NM_001243014
NM_019624
NM_019625
NM_203444
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9
|
|
chr15_-_37390372
|
2.971
|
NM_172315
|
MEIS2
|
Meis homeobox 2
|
|
chr15_+_41221530
|
2.952
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr2_+_43864433
|
2.948
|
NM_172069
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2
|
|
chr10_-_21463019
|
2.911
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr1_-_225840660
|
2.911
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr12_-_88974094
|
2.903
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr12_-_71031174
|
2.813
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr11_+_19372270
|
2.804
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr1_+_215256559
|
2.792
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr9_+_137533643
|
2.773
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr8_+_134203281
|
2.729
|
NM_001204869
NM_001204870
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr15_+_96869156
|
2.710
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_+_11034419
|
2.697
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr10_+_18549583
|
2.688
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr18_-_30050394
|
2.668
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr13_+_113623496
|
2.655
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr6_-_88854989
|
2.651
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr5_-_179780314
|
2.640
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr10_+_18689512
|
2.638
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr21_+_18885104
|
2.633
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr6_-_99797530
|
2.624
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chrX_-_54384436
|
2.621
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr11_+_34654010
|
2.616
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr6_-_56112315
|
2.615
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr15_-_37391596
|
2.612
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr2_+_210636700
|
2.596
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr15_+_96876568
|
2.585
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr14_+_67999915
|
2.553
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr16_+_66878281
|
2.535
|
NM_005182
|
CA7
|
carbonic anhydrase VII
|
|
chr5_+_172068188
|
2.531
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr4_+_55095263
|
2.529
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr21_-_40033617
|
2.521
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_81991959
|
2.499
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr1_+_205012329
|
2.483
|
NM_005076
|
CNTN2
|
contactin 2 (axonal)
|
|
chr9_-_140196702
|
2.459
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr13_-_45775174
|
2.458
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr1_-_205325915
|
2.444
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr12_-_81763127
|
2.415
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr5_-_121414011
|
2.414
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr9_+_112403067
|
2.387
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr1_-_117210301
|
2.372
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr15_-_63674074
|
2.349
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr13_-_103054027
|
2.329
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr3_+_183993798
|
2.326
|
NM_001037324
NM_001100120
NM_001100121
|
ECE2
|
endothelin converting enzyme 2
|
|
chr10_-_21186530
|
2.322
|
NM_006393
|
NEBL
|
nebulette
|
|
chr10_-_21786205
|
2.309
|
NM_001010911
|
C10orf114
|
chromosome 10 open reading frame 114
|
|
chr19_+_19322763
|
2.306
|
NM_004386
|
NCAN
|
neurocan
|
|
chr12_-_82152269
|
2.290
|
NM_001220475
NM_001220476
NM_001220473
NM_001220474
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr12_-_82153092
|
2.276
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr13_-_102568994
|
2.264
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr18_+_20715726
|
2.245
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chrX_+_102631267
|
2.199
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr8_-_10587942
|
2.180
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr4_-_139163222
|
2.176
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr6_-_119399759
|
2.174
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr1_-_166944504
|
2.161
|
NM_199351
|
ILDR2
|
immunoglobulin-like domain containing receptor 2
|
|
chr6_+_44238479
|
2.150
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr9_-_14398981
|
2.133
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr13_-_80915041
|
2.130
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr7_-_123673486
|
2.115
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr10_-_113943467
|
2.105
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr1_-_75139421
|
2.075
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr9_-_93405066
|
2.075
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr19_-_2721337
|
2.064
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr12_-_105478218
|
2.054
|
NM_001034173
|
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2
|
|
chr22_+_41957011
|
2.051
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr9_+_103947316
|
2.050
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr1_+_65613771
|
2.040
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr1_+_215178884
|
2.040
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr21_+_33245304
|
2.025
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr6_+_142622797
|
2.019
|
NM_001032394
NM_001032395
NM_020455
NM_198569
|
GPR126
|
G protein-coupled receptor 126
|
|
chr18_-_31803514
|
1.998
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr6_+_13925097
|
1.995
|
NM_001165032
NM_152737
|
RNF182
|
ring finger protein 182
|
|
chr7_+_127881330
|
1.990
|
NM_000230
|
LEP
|
leptin
|
|
chr12_+_12938540
|
1.990
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr11_-_45687135
|
1.986
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr1_+_117452359
|
1.975
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr18_+_20735788
|
1.973
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr17_-_41985112
|
1.958
|
NM_005374
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr9_-_14314036
|
1.956
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr17_+_36584692
|
1.926
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr6_-_153452383
|
1.925
|
NM_012419
|
RGS17
|
regulator of G-protein signaling 17
|
|
chr1_-_99470177
|
1.923
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr5_+_152870083
|
1.923
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr20_-_14318200
|
1.920
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr1_-_35370983
|
1.914
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr1_+_205473683
|
1.909
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr16_+_66878840
|
1.899
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr22_-_37098620
|
1.897
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr1_+_65613211
|
1.894
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr18_-_812268
|
1.886
|
NM_005433
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1
|
|
chr22_-_29075708
|
1.883
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr1_+_204913353
|
1.874
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr8_+_77593506
|
1.857
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr16_+_1756161
|
1.844
|
NM_001040439
NM_015133
|
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr8_+_28351694
|
1.833
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr10_+_18629613
|
1.815
|
NM_201590
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr3_+_12330336
|
1.806
|
NM_138711
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chrX_+_102632108
|
1.803
|
NM_014380
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr7_-_27239724
|
1.793
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr6_-_3445198
|
1.792
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr20_-_41818464
|
1.787
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr6_+_13925423
|
1.780
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr8_+_16884745
|
1.779
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr14_-_30396811
|
1.768
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr1_+_120254418
|
1.765
|
NM_006623
|
PHGDH
|
phosphoglycerate dehydrogenase
|
|
chr18_+_49866541
|
1.761
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr1_+_13910251
|
1.761
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr19_+_57154526
|
1.756
|
NM_001193628
|
LOC147670
|
uncharacterized LOC147670
|
|
chr12_-_71003622
|
1.754
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr2_-_20212448
|
1.746
|
NM_002381
|
MATN3
|
matrilin 3
|
|
chr4_+_113739203
|
1.744
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr6_+_148663728
|
1.742
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr8_-_97172943
|
1.734
|
NM_001001557
|
GDF6
|
growth differentiation factor 6
|
|
chr12_+_5019048
|
1.726
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr11_-_88796815
|
1.719
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr19_+_1285766
|
1.716
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr18_-_31802433
|
1.708
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr6_-_46620229
|
1.708
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr3_+_12329332
|
1.705
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr4_-_177713664
|
1.701
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chr7_+_98246595
|
1.694
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr9_-_101470836
|
1.690
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr9_+_34958171
|
1.686
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chr17_+_42385780
|
1.679
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr3_+_150804675
|
1.674
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr12_+_54955226
|
1.659
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr15_+_54305100
|
1.651
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr14_+_37667116
|
1.650
|
NM_001195296
NM_001195297
NM_138731
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr18_+_55102777
|
1.649
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|