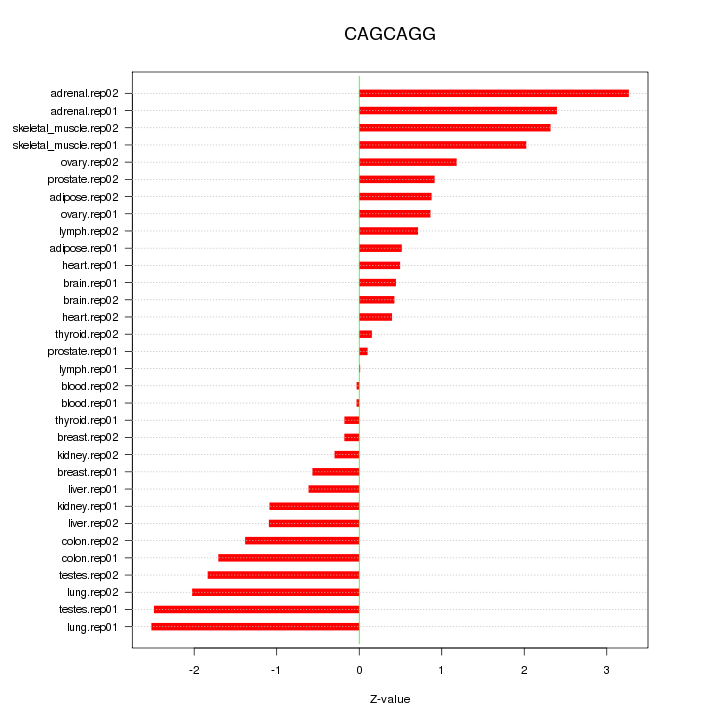
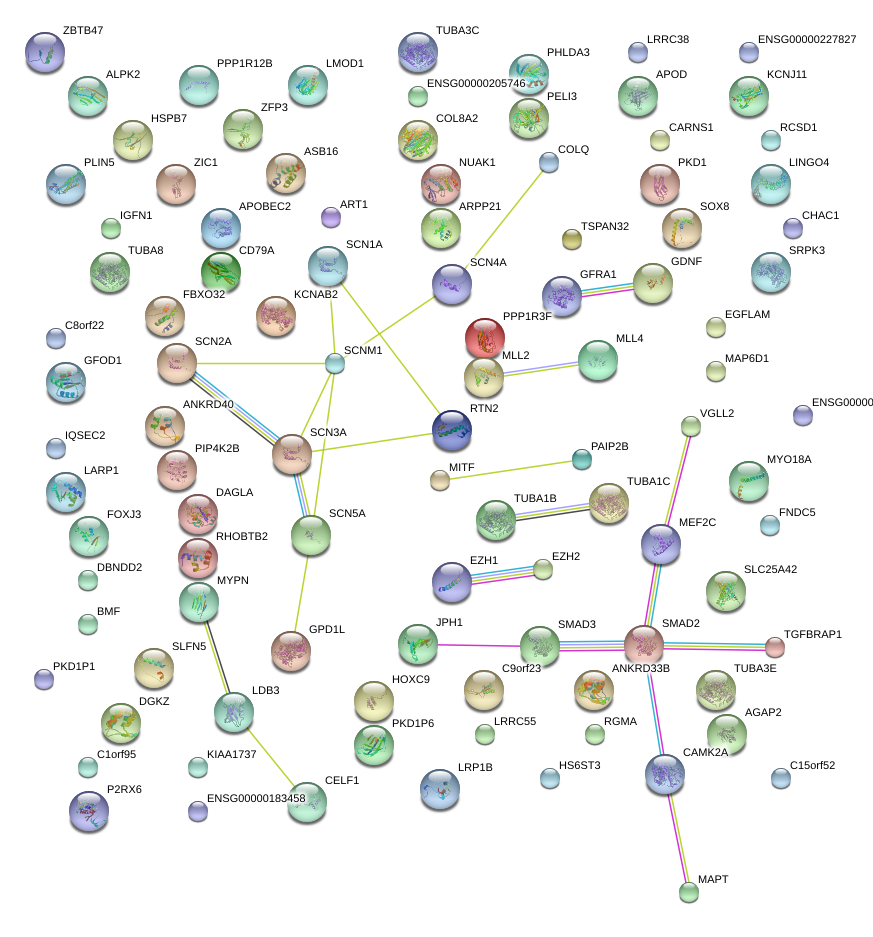
|
chr11_-_17410205
|
2.326
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr11_-_17410863
|
2.186
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr8_-_124553448
|
2.068
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr8_-_124544376
|
2.041
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr1_-_16345284
|
1.708
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr18_-_56296188
|
1.677
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr11_+_66234293
|
1.495
|
NM_001098510
NM_001243136
NM_145065
|
PELI3
|
pellino homolog 3 (Drosophila)
|
|
chr8_-_75233461
|
1.478
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr22_+_21369441
|
1.404
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr1_-_33338081
|
1.404
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr5_-_149669400
|
1.379
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr17_+_43971655
|
1.375
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr17_+_42248073
|
1.338
|
NM_080863
|
ASB16
|
ankyrin repeat and SOCS box containing 16
|
|
chr8_+_49984902
|
1.322
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr11_+_67183144
|
1.297
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr15_+_41245635
|
1.263
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr11_+_3666357
|
1.251
|
NM_004314
|
ART1
|
ADP-ribosyltransferase 1
|
|
chr1_-_151777741
|
1.202
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr19_-_45996470
|
1.200
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chr10_+_69869249
|
1.200
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_+_202431870
|
1.196
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr7_+_140774031
|
1.189
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr6_+_41020939
|
1.182
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr3_+_42695175
|
1.170
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr20_+_44036628
|
1.141
|
NM_001048225
NM_001048226
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr19_-_46000159
|
1.106
|
NM_005619
NM_206900
|
RTN2
|
reticulon 2
|
|
chr3_-_183543295
|
1.105
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr20_+_44034632
|
1.100
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr1_+_202509241
|
1.096
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr15_-_93617091
|
1.094
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr5_+_10564431
|
1.091
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr15_-_93616891
|
1.079
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr3_-_195310985
|
1.077
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr15_-_93632161
|
1.075
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr19_+_42381189
|
1.073
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr12_-_49449106
|
1.059
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr17_+_4981721
|
1.057
|
NM_153018
|
ZFP3
|
zinc finger protein 3 homolog (mouse)
|
|
chr11_+_46383115
|
1.040
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr17_-_36956145
|
1.025
|
NM_003559
|
PIP4K2B
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr12_-_58135853
|
1.011
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr14_+_77564570
|
1.011
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr6_+_117586720
|
1.003
|
NM_153453
NM_182645
|
VGLL2
|
vestigial like 2 (Drosophila)
|
|
chr17_-_48785016
|
1.000
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr12_+_54393876
|
0.995
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr17_-_27507195
|
0.993
|
NM_078471
NM_203318
|
MYO18A
|
myosin XVIIIA
|
|
chr15_-_40633122
|
0.993
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr15_-_93616358
|
0.987
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr3_+_69812620
|
0.981
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_+_56949220
|
0.950
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr1_-_201915715
|
0.929
|
NM_012134
|
LMOD1
|
leiomodin 1 (smooth muscle)
|
|
chr13_+_96743092
|
0.914
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr20_+_44035203
|
0.913
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr3_+_69812961
|
0.899
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr17_-_40897025
|
0.898
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr15_-_93631751
|
0.897
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr5_-_88199868
|
0.895
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr1_+_167599329
|
0.891
|
NM_052862
|
RCSD1
|
RCSD domain containing 1
|
|
chr11_-_47510518
|
0.883
|
NM_001025596
NM_001172640
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr3_+_32148000
|
0.881
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chrX_+_153046422
|
0.874
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr1_+_201159952
|
0.866
|
NM_001164586
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
|
chrX_-_53310748
|
0.862
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr5_+_154092461
|
0.861
|
NM_015315
|
LARP1
|
La ribonucleoprotein domain family, member 1
|
|
chr1_-_13840157
|
0.845
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr15_+_67418053
|
0.842
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_88119604
|
0.841
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr15_-_40398286
|
0.836
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chr11_+_61447816
|
0.832
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chrX_+_49126299
|
0.820
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr2_-_142889269
|
0.817
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr22_+_18593399
|
0.810
|
NM_001193414
NM_018943
|
TUBA8
|
tubulin, alpha 8
|
|
chr17_+_33570079
|
0.808
|
NM_144975
|
SLFN5
|
schlafen family member 5
|
|
chr2_-_71454200
|
0.806
|
NM_020459
|
PAIP2B
|
poly(A) binding protein interacting protein 2B
|
|
chr1_-_33336289
|
0.805
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr6_-_13408368
|
0.797
|
NM_001242630
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr5_+_38445577
|
0.796
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_-_201438150
|
0.792
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr3_-_15563160
|
0.791
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr16_-_2185824
|
0.789
|
NM_000296
NM_001009944
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant)
|
|
chr11_-_47516072
|
0.788
|
NM_198700
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr2_-_105946104
|
0.786
|
NM_001142621
NM_004257
|
TGFBRAP1
|
transforming growth factor, beta receptor associated protein 1
|
|
chr2_-_166060552
|
0.778
|
NM_001081676
NM_001081677
NM_006922
|
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit
|
|
chr11_+_46354454
|
0.774
|
NM_201532
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr3_-_38691118
|
0.774
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr1_+_151138497
|
0.771
|
NM_001204856
NM_024041
|
SCNM1
|
sodium channel modifier 1
|
|
chr11_+_46366816
|
0.769
|
NM_201533
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr3_+_147127151
|
0.765
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr6_+_53883713
|
0.757
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr1_-_42801529
|
0.752
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chr10_+_88428167
|
0.740
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr1_+_6105980
|
0.738
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_+_46368881
|
0.732
|
NM_001199266
NM_001199267
NM_001199268
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr1_+_226736500
|
0.728
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr16_+_1031807
|
0.726
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr15_-_40398573
|
0.726
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr1_-_36565849
|
0.722
|
NM_005202
|
COL8A2
|
collagen, type VIII, alpha 2
|
|
chr9_-_34612075
|
0.717
|
NM_148178
NM_148179
|
C9orf23
|
chromosome 9 open reading frame 23
|
|
chr3_+_69788562
|
0.714
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_-_15540378
|
0.714
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr5_-_37839781
|
0.710
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr19_+_19174792
|
0.708
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr5_+_38403633
|
0.707
|
NM_182798
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_+_6106173
|
0.703
|
NM_001199863
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_-_47545539
|
0.701
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr3_+_35721115
|
0.700
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_-_106533810
|
0.698
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr19_-_4535202
|
0.698
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr8_+_22844918
|
0.698
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr10_-_118032717
|
0.694
|
NM_001145453
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr12_+_122516555
|
0.693
|
NM_014938
|
MLXIP
|
MLX interacting protein
|
|
chr20_-_42816217
|
0.692
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chrX_+_135251454
|
0.690
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr11_-_73693883
|
0.690
|
NM_003355
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier)
|
|
chr14_+_90863302
|
0.685
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr9_-_140444835
|
0.682
|
NM_001098537
NM_152286
|
PNPLA7
|
patatin-like phospholipase domain containing 7
|
|
chr5_-_88179278
|
0.679
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_35696359
|
0.676
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_-_33647626
|
0.669
|
NM_018207
|
TRIM62
|
tripartite motif containing 62
|
|
chr20_+_55966440
|
0.669
|
NM_017495
NM_183425
|
RBM38
|
RNA binding motif protein 38
|
|
chr9_-_134585107
|
0.668
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr15_-_40401041
|
0.668
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr5_-_37835928
|
0.662
|
NM_001190468
NM_001190469
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr9_-_134612924
|
0.652
|
NM_005312
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr10_-_118032975
|
0.652
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr3_+_141043054
|
0.652
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr6_-_39895317
|
0.651
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr14_+_60712469
|
0.651
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr3_-_44519156
|
0.651
|
NM_181489
|
ZNF445
|
zinc finger protein 445
|
|
chr15_+_91643429
|
0.650
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr4_-_42658883
|
0.649
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr1_+_210001267
|
0.648
|
NM_014388
|
DIEXF
|
digestive organ expansion factor homolog (zebrafish)
|
|
chr3_+_50654558
|
0.643
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr9_-_124989755
|
0.642
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr8_+_22853360
|
0.642
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr17_-_62050200
|
0.636
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr6_-_13486382
|
0.636
|
NM_001242628
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr5_+_155753755
|
0.635
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr4_+_3076236
|
0.630
|
NM_002111
|
HTT
|
huntingtin
|
|
chr5_-_37835592
|
0.626
|
NM_199231
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr1_+_145611018
|
0.621
|
NM_014455
|
RNF115
|
ring finger protein 115
|
|
chr17_+_72244504
|
0.618
|
NM_052869
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr1_+_6086072
|
0.618
|
NM_003636
NM_172130
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr7_+_128506463
|
0.615
|
NM_001195150
|
LOC100130705
|
uncharacterized LOC100130705
|
|
chr17_+_72209500
|
0.613
|
NM_032646
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr4_-_53522756
|
0.612
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr8_-_93029864
|
0.612
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_58132028
|
0.608
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr11_-_71791434
|
0.608
|
NM_006185
|
NUMA1
|
nuclear mitotic apparatus protein 1
|
|
chr16_-_90076528
|
0.607
|
NM_024043
|
DBNDD1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1
|
|
chr5_+_176560056
|
0.605
|
NM_172349
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr15_+_65369153
|
0.600
|
NM_001101362
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13
|
|
chr20_+_56964162
|
0.597
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr10_+_16478941
|
0.596
|
NM_001001484
NM_030664
|
PTER
|
phosphotriesterase related
|
|
chr17_+_78518611
|
0.588
|
NM_001163034
NM_020761
|
RPTOR
|
regulatory associated protein of MTOR, complex 1
|
|
chr1_-_154832187
|
0.582
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr16_-_73082169
|
0.582
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr15_-_64338519
|
0.581
|
NM_014326
|
DAPK2
|
death-associated protein kinase 2
|
|
chr22_-_44893903
|
0.579
|
NM_032287
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr7_+_150065859
|
0.579
|
NM_001099695
NM_001099696
NM_013400
|
REPIN1
|
replication initiator 1
|
|
chr19_-_48867149
|
0.579
|
NM_018273
|
TMEM143
|
transmembrane protein 143
|
|
chr19_-_49222956
|
0.575
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr1_-_42800883
|
0.568
|
NM_001198852
|
FOXJ3
|
forkhead box J3
|
|
chr17_-_8055684
|
0.565
|
NM_002616
|
PER1
|
period homolog 1 (Drosophila)
|
|
chr15_-_42749710
|
0.565
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr17_+_1182956
|
0.564
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr1_+_6052357
|
0.563
|
NM_001199861
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr3_-_119812973
|
0.563
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr15_+_91643181
|
0.558
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr11_+_66024746
|
0.556
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr1_-_55681024
|
0.556
|
NM_015306
|
USP24
|
ubiquitin specific peptidase 24
|
|
chr17_-_16875388
|
0.555
|
NM_012452
|
TNFRSF13B
|
tumor necrosis factor receptor superfamily, member 13B
|
|
chr3_+_69915374
|
0.554
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_-_53164023
|
0.553
|
NM_023077
|
SELRC1
|
Sel1 repeat containing 1
|
|
chr22_-_30752880
|
0.553
|
NM_001005409
NM_005877
|
SF3A1
|
splicing factor 3a, subunit 1, 120kDa
|
|
chr15_+_85359910
|
0.552
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr3_+_35722428
|
0.550
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr6_-_32977351
|
0.549
|
NM_002119
|
HLA-DOA
|
major histocompatibility complex, class II, DO alpha
|
|
chr11_-_1593149
|
0.549
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr7_+_4721722
|
0.548
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr14_-_75389144
|
0.546
|
NM_031464
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr1_-_32403975
|
0.546
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chrX_+_135251795
|
0.543
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_-_142054431
|
0.543
|
NM_020724
|
RNF150
|
ring finger protein 150
|
|
chr1_-_207095198
|
0.542
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr19_-_49220213
|
0.539
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr21_-_43346780
|
0.538
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr3_+_69985750
|
0.537
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_-_42800902
|
0.531
|
NM_014947
|
FOXJ3
|
forkhead box J3
|
|
chr1_-_154842725
|
0.531
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr11_+_64781584
|
0.526
|
NM_001199745
NM_001667
|
ARL2-SNX15
ARL2
|
ARL2-SNX15 readthrough
ADP-ribosylation factor-like 2
|
|
chr15_-_56535475
|
0.526
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr1_-_33841142
|
0.525
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chrX_+_135278912
|
0.524
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr18_+_11689135
|
0.523
|
NM_182978
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type
|
|
chr1_+_6094342
|
0.522
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chrX_-_77041678
|
0.522
|
NM_000489
NM_138270
|
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked
|
|
chr19_+_6739697
|
0.521
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr15_+_67458492
|
0.518
|
NM_001145104
|
SMAD3
|
SMAD family member 3
|
|
chr3_+_151985828
|
0.518
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr17_+_1619816
|
0.517
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|