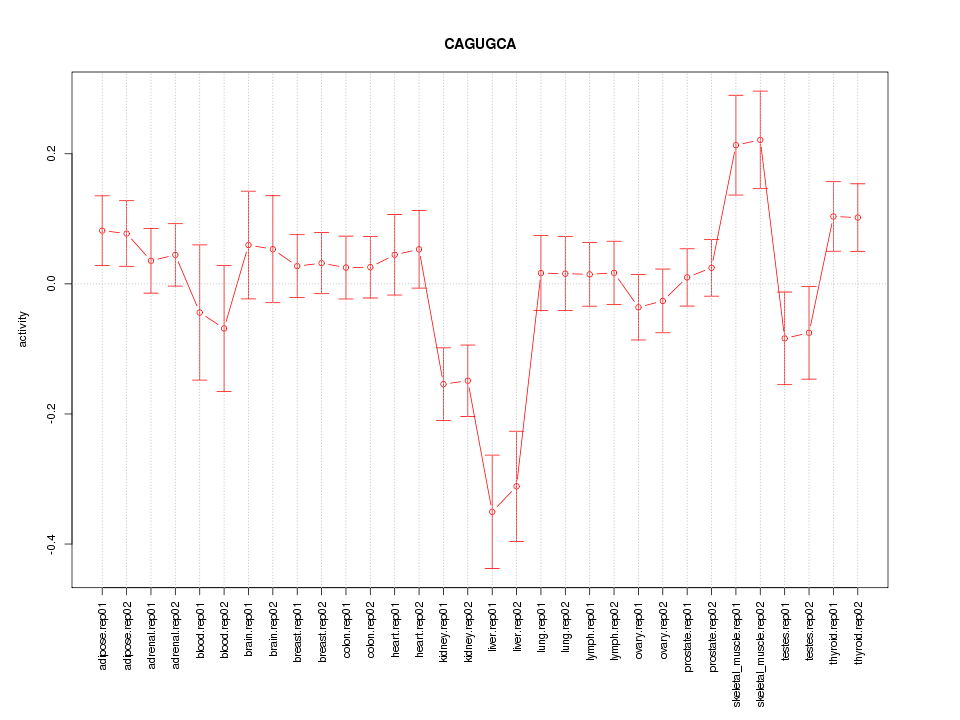
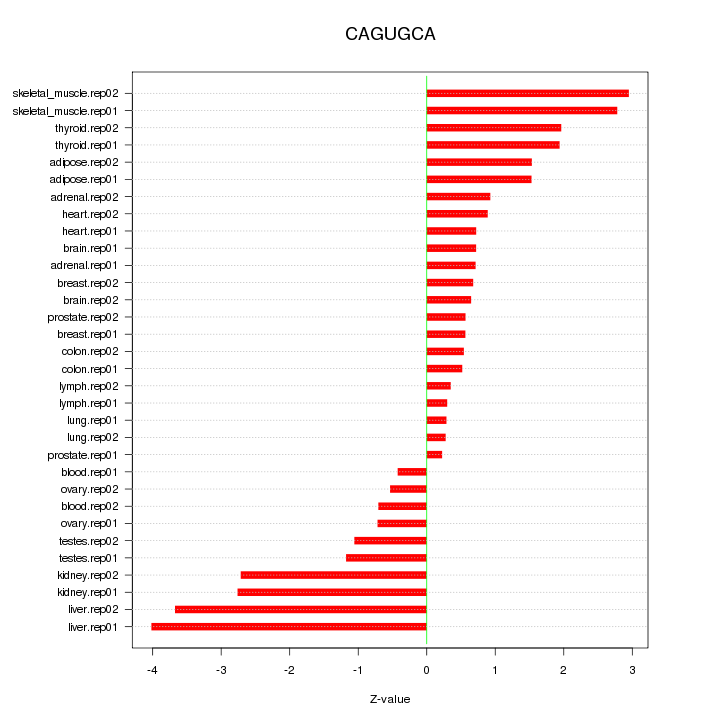
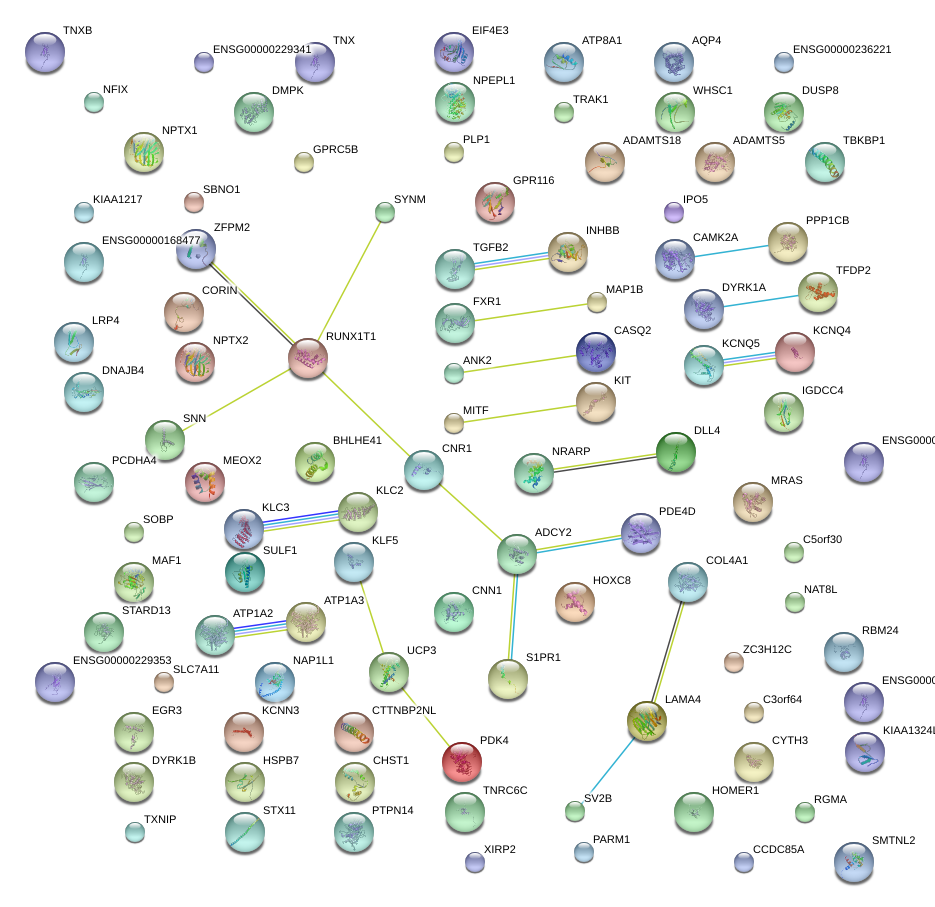
|
chr5_+_7396030
|
5.597
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr6_+_17282931
|
5.580
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr5_-_149669400
|
5.298
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr6_+_17281773
|
5.091
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr12_-_26277807
|
4.954
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr15_-_93632161
|
4.848
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616891
|
4.597
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr15_-_93616358
|
4.588
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr6_+_17282294
|
4.512
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr15_-_93617091
|
4.303
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr1_+_160085519
|
4.257
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr3_+_69812620
|
4.006
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_-_93631751
|
3.857
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr6_+_73331570
|
3.778
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr3_+_69812961
|
3.757
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr18_-_24445652
|
3.516
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr7_-_15726273
|
3.314
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr13_-_33760215
|
3.233
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr2_+_121103670
|
3.188
|
NM_002193
|
INHBB
|
inhibin, beta B
|
|
chr3_+_138066489
|
3.186
|
NM_001252091
NM_012219
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr3_+_69788562
|
3.182
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_218518675
|
3.007
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr6_-_88854989
|
2.973
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr18_-_24442534
|
2.932
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr21_-_28339405
|
2.875
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr19_-_46283763
|
2.867
|
NM_001081563
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr19_-_46285752
|
2.750
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr16_+_11762235
|
2.719
|
NM_003498
|
SNN
|
stannin
|
|
chr4_-_42658883
|
2.676
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr2_+_167744996
|
2.641
|
NM_001079810
NM_001199143
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr7_-_86595203
|
2.612
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr6_-_88875766
|
2.579
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr3_+_138067444
|
2.546
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr5_-_58334936
|
2.520
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_41221530
|
2.490
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr11_-_46939908
|
2.489
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr5_-_58882274
|
2.484
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_95225802
|
2.443
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr6_-_32076997
|
2.422
|
NM_019105
|
TNXB
|
tenascin XB
|
|
chr11_-_73720281
|
2.402
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr12_-_123834984
|
2.396
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr5_-_58571916
|
2.376
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_59783885
|
2.328
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_69915374
|
2.324
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_145438437
|
2.304
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr11_-_1593149
|
2.285
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr12_+_54402795
|
2.281
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr5_-_59064437
|
2.280
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_93029864
|
2.184
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_-_140196702
|
2.164
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr10_+_24497719
|
2.112
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr15_-_65715385
|
2.111
|
NM_020962
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4
|
|
chr20_+_57264186
|
2.085
|
NM_001204872
NM_001204873
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr3_-_141719188
|
2.064
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr19_+_11649578
|
2.031
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr5_-_59189620
|
2.027
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_78450247
|
2.012
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr3_+_42132707
|
2.007
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr2_+_168043792
|
2.006
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr5_-_78809658
|
2.003
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr1_-_154832187
|
1.975
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr5_-_58295758
|
1.946
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_+_13105970
|
1.918
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr3_-_71803923
|
1.913
|
NM_173359
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr3_+_69985750
|
1.860
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_139163222
|
1.850
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr11_-_45687135
|
1.843
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr8_+_70405027
|
1.824
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr8_+_106331146
|
1.815
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr3_+_180630054
|
1.814
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr7_-_6312137
|
1.804
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr1_-_16345284
|
1.804
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr1_+_101702304
|
1.778
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chrX_+_103031433
|
1.771
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr4_+_113739203
|
1.760
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr1_+_112938799
|
1.754
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr6_-_112575766
|
1.750
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr4_+_1894508
|
1.745
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr10_+_23983674
|
1.704
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr4_+_75858205
|
1.687
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr5_-_58652671
|
1.683
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr13_-_110959441
|
1.671
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr3_-_71803513
|
1.668
|
NM_001134649
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr16_-_19896105
|
1.666
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr1_-_154842725
|
1.665
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chrX_+_103031753
|
1.659
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr19_-_40324747
|
1.658
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr1_-_214724974
|
1.657
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr11_+_109964086
|
1.655
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr5_+_102594406
|
1.637
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr2_+_56411123
|
1.634
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr15_+_99645152
|
1.630
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr1_+_78470593
|
1.580
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr13_+_98605928
|
1.576
|
NM_002271
|
IPO5
|
importin 5
|
|
chr20_+_57267818
|
1.572
|
NM_024663
|
NPEPL1
|
aminopeptidase-like 1
|
|
chr4_+_113970784
|
1.542
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr16_-_77468930
|
1.505
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr7_+_98246595
|
1.498
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr8_+_145159304
|
1.480
|
NM_032272
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr15_+_91643429
|
1.480
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr7_-_86688944
|
1.463
|
NM_001142749
|
KIAA1324L
|
KIAA1324-like
|
|
chr1_-_116311398
|
1.439
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr8_-_22550708
|
1.405
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr17_+_4487275
|
1.396
|
NM_198501
|
SMTNL2
|
smoothelin-like 2
|
|
chr6_+_144471631
|
1.393
|
NM_003764
|
STX11
|
syntaxin 11
|
|
chr6_+_107811272
|
1.388
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr4_+_55523906
|
1.380
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr17_+_76000248
|
1.377
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr12_-_76478737
|
1.364
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr5_+_71403040
|
1.358
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_71778268
|
1.337
|
NM_001134650
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr2_+_28974608
|
1.326
|
NM_206876
NM_002709
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme
|
|
chr6_-_46922533
|
1.321
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr11_+_66024746
|
1.317
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr3_-_69062744
|
1.317
|
NM_173654
|
C3orf64
|
chromosome 3 open reading frame 64
|
|
chr13_+_73633097
|
1.296
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr17_+_45772629
|
1.296
|
NM_014726
|
TBKBP1
|
TBK1 binding protein 1
|
|
chr4_+_2061238
|
1.281
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr21_+_38791206
|
1.281
|
NM_130436
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr14_-_61190458
|
1.278
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr3_-_71774407
|
1.277
|
NM_001134651
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr17_+_4487833
|
1.268
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr12_-_8088629
|
1.267
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr14_-_27066959
|
1.258
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr18_-_53255734
|
1.245
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr8_-_22550057
|
1.241
|
NM_001199881
|
EGR3
|
early growth response 3
|
|
chr11_+_66025070
|
1.240
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr18_-_12656730
|
1.240
|
NM_001128627
|
SPIRE1
|
spire homolog 1 (Drosophila)
|
|
chr1_-_51887740
|
1.239
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr4_-_120549115
|
1.235
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr15_+_91643181
|
1.235
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr8_-_22549901
|
1.228
|
NM_001199880
|
EGR3
|
early growth response 3
|
|
chr4_-_120549827
|
1.227
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr3_+_2933898
|
1.226
|
NM_001206956
NM_175613
|
CNTN4
|
contactin 4
|
|
chr18_+_43753986
|
1.224
|
NM_001008239
NM_145055
|
C18orf25
|
chromosome 18 open reading frame 25
|
|
chr11_+_6280903
|
1.224
|
NM_176875
|
CCKBR
|
cholecystokinin B receptor
|
|
chr8_-_93107881
|
1.224
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_70378858
|
1.213
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr4_-_120548329
|
1.209
|
NM_033430
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr11_+_112831968
|
1.205
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr9_+_100263461
|
1.203
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr12_-_50101192
|
1.202
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr1_+_11249344
|
1.200
|
NM_021146
|
ANGPTL7
|
angiopoietin-like 7
|
|
chr3_+_2280512
|
1.185
|
NM_001206955
|
CNTN4
|
contactin 4
|
|
chr11_-_62446395
|
1.177
|
NM_015853
|
UBXN1
|
UBX domain protein 1
|
|
chr1_-_46598250
|
1.175
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr12_+_26126687
|
1.173
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_179112178
|
1.165
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr1_+_204485461
|
1.155
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr17_-_7232455
|
1.147
|
NM_001005408
NM_032442
|
NEURL4
|
neuralized homolog 4 (Drosophila)
|
|
chr1_-_95007139
|
1.137
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr5_-_126366439
|
1.135
|
NM_178450
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3
|
|
chr3_-_52479042
|
1.128
|
NM_020163
|
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr21_+_38792600
|
1.121
|
NM_130438
NM_001396
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr4_+_1873035
|
1.119
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr7_-_87856282
|
1.117
|
NM_198901
|
SRI
|
sorcin
|
|
chr16_-_30107492
|
1.109
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr5_-_142065991
|
1.104
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr15_-_48937795
|
1.100
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr3_+_187943192
|
1.098
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_+_19140674
|
1.092
|
NM_001102664
NM_014964
NM_148921
|
EPN2
|
epsin 2
|
|
chr3_+_127641901
|
1.080
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr20_+_34700257
|
1.079
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr5_-_138775213
|
1.073
|
NM_152686
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18
|
|
chr9_-_89562103
|
1.064
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr6_-_46889620
|
1.058
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr17_+_47865980
|
1.058
|
NM_001199155
NM_001199156
NM_001199157
NM_001199158
NM_007067
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr17_-_66287256
|
1.046
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr10_+_102295572
|
1.041
|
NM_017902
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor
|
|
chr10_-_131762017
|
1.032
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr7_-_107204705
|
1.028
|
NM_001161520
NM_006348
NM_181733
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr11_-_118016134
|
1.028
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr20_+_34359904
|
1.013
|
NM_016436
|
PHF20
|
PHD finger protein 20
|
|
chr2_-_238322840
|
1.000
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_+_33004771
|
0.998
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr1_+_202431870
|
0.995
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr11_-_118023534
|
0.994
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr11_+_125034558
|
0.985
|
NM_022062
|
PKNOX2
|
PBX/knotted 1 homeobox 2
|
|
chr3_+_42695175
|
0.982
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chrX_-_43832920
|
0.981
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr5_+_95997723
|
0.979
|
NM_001190442
|
CAST
|
calpastatin
|
|
chr15_+_51200864
|
0.978
|
NM_001252127
NM_007347
|
AP4E1
|
adaptor-related protein complex 4, epsilon 1 subunit
|
|
chr5_-_142077508
|
0.975
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_115342150
|
0.968
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr5_+_95997871
|
0.965
|
NM_001042440
|
CAST
|
calpastatin
|
|
chr18_+_32397925
|
0.961
|
NM_001198943
|
DTNA
|
dystrobrevin, alpha
|
|
chrX_-_19988381
|
0.960
|
NM_198279
|
CXorf23
|
chromosome X open reading frame 23
|
|
chr1_+_202509241
|
0.959
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr17_+_54671059
|
0.957
|
NM_005450
|
NOG
|
noggin
|
|
chr18_+_32398225
|
0.957
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr1_-_109584589
|
0.951
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr10_+_60094712
|
0.938
|
NM_001204880
NM_003338
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1
|
|
chr8_-_93107442
|
0.935
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_-_118047336
|
0.930
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr3_+_2140449
|
0.915
|
NM_175607
|
CNTN4
|
contactin 4
|
|
chr12_-_110505499
|
0.912
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr2_-_9695885
|
0.909
|
NM_003183
|
ADAM17
|
ADAM metallopeptidase domain 17
|
|
chr18_+_7567313
|
0.908
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr17_+_80477564
|
0.904
|
NM_004514
|
FOXK2
|
forkhead box K2
|
|
chr7_+_116140097
|
0.900
|
NM_001206748
|
CAV2
|
caveolin 2
|