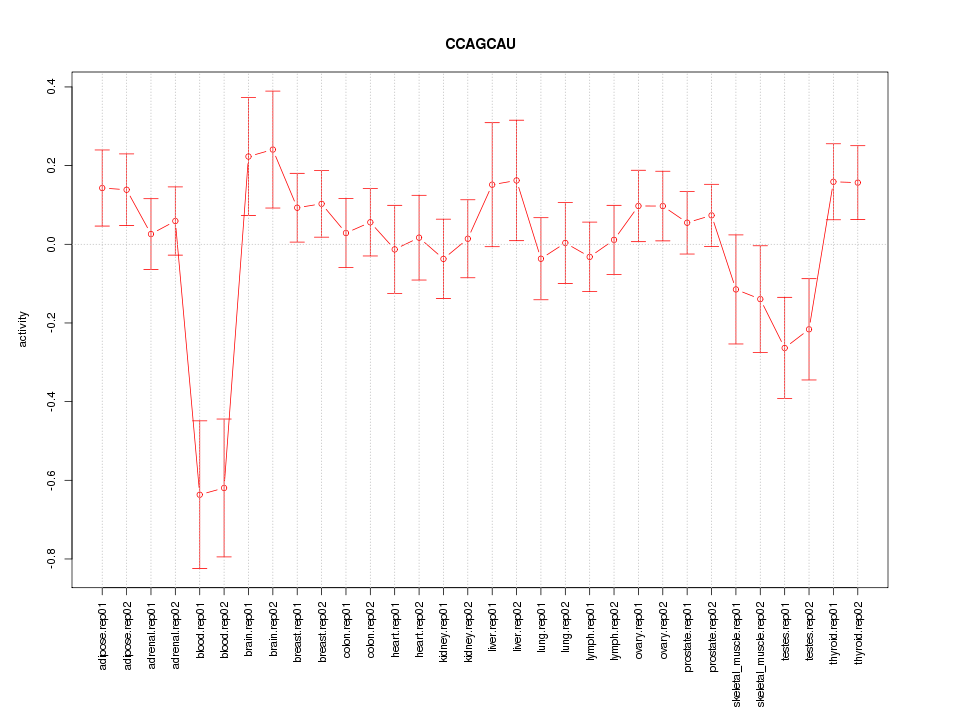
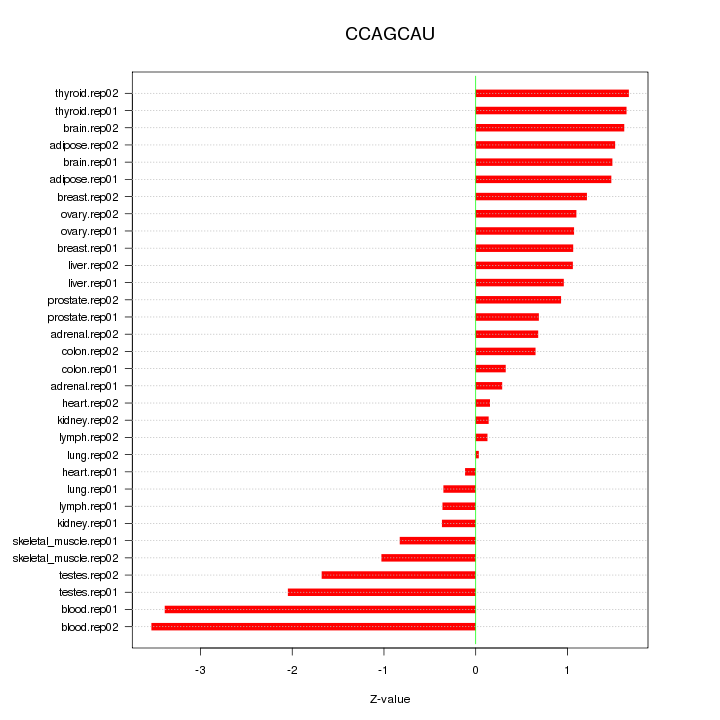
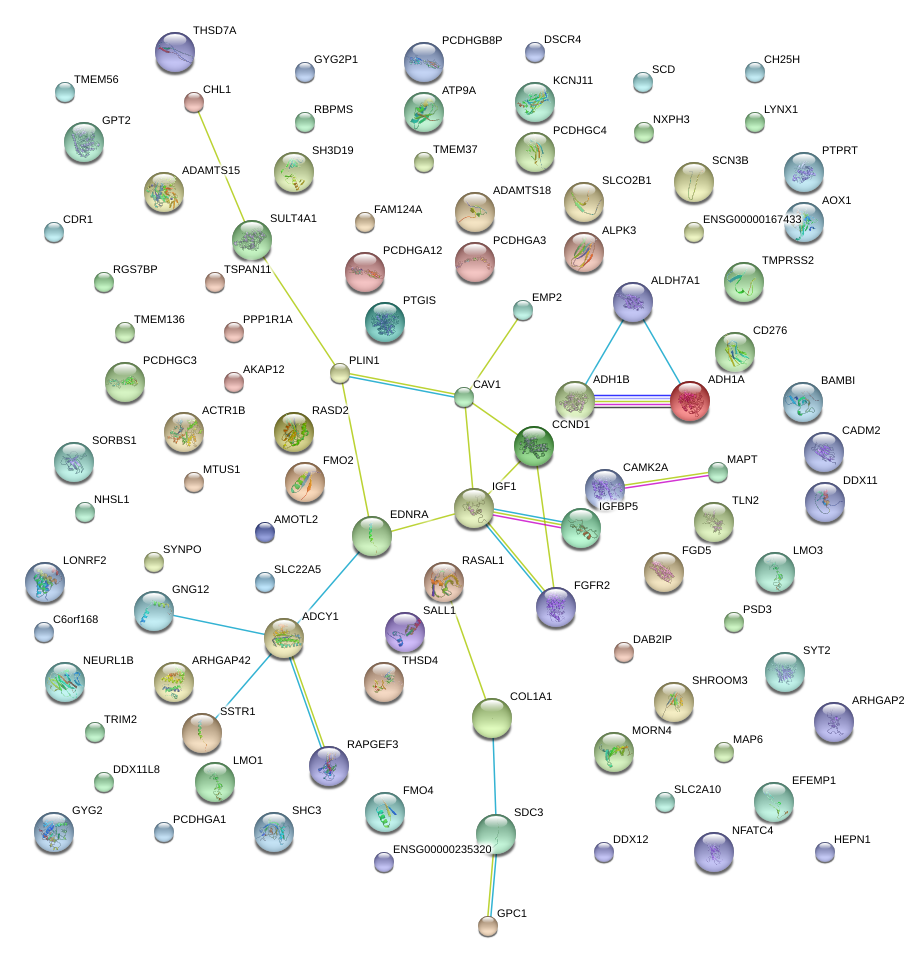
|
chr12_-_54982271
|
4.195
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr3_-_134093235
|
4.138
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr11_+_130318457
|
4.057
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr20_+_45338125
|
4.027
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr3_+_85008031
|
4.019
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr16_-_51184507
|
3.827
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr9_+_124329398
|
3.806
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr16_-_51185150
|
3.675
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr3_+_238274
|
3.247
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr22_-_44258210
|
3.175
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr15_+_85359910
|
3.167
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr10_+_102106771
|
3.164
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr7_+_116166346
|
3.094
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr10_-_123357503
|
2.941
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr15_+_73976621
|
2.880
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|
|
chr3_+_85775631
|
2.846
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr8_-_18871163
|
2.793
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr15_+_62939509
|
2.672
|
NM_015059
|
TLN2
|
talin 2
|
|
chr7_+_116164838
|
2.603
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr17_-_48278987
|
2.570
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr5_+_140723600
|
2.566
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr10_-_123356158
|
2.530
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr21_-_42879908
|
2.527
|
NM_001135099
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr5_+_140718353
|
2.523
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr7_+_116165043
|
2.496
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr5_+_140753480
|
2.482
|
NM_018919
NM_032086
|
PCDHGA6
|
protocadherin gamma subfamily A, 6
|
|
chr10_+_28966419
|
2.456
|
NM_012342
|
BAMBI
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis)
|
|
chr8_-_17658385
|
2.448
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr5_+_140782367
|
2.369
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr11_+_100557851
|
2.312
|
NM_152432
|
ARHGAP42
|
Rho GTPase activating protein 42
|
|
chr10_-_123353771
|
2.311
|
NM_001144916
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr5_+_140739589
|
2.251
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr8_-_18666295
|
2.224
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr5_+_140864626
|
2.214
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr2_+_120189342
|
2.123
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr1_+_95558072
|
2.093
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr8_-_17580024
|
2.092
|
NM_001166393
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chrX_-_139866582
|
2.055
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chr16_+_46918216
|
1.982
|
NM_133443
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2
|
|
chr12_-_113573267
|
1.976
|
NM_001193521
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr2_-_100939194
|
1.969
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr21_-_42880080
|
1.931
|
NM_005656
|
TMPRSS2
|
transmembrane protease, serine 2
|
|
chr8_-_17579729
|
1.928
|
NM_001001931
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr11_-_75379414
|
1.918
|
NM_033063
NM_207577
|
MAP6
|
microtubule-associated protein 6
|
|
chr8_-_143858436
|
1.905
|
NM_177476
NM_177477
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr5_+_140729723
|
1.904
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr13_+_51796363
|
1.897
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr8_-_17555158
|
1.892
|
NM_020749
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr10_-_123353330
|
1.891
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr5_+_140749830
|
1.891
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr12_+_31079837
|
1.887
|
NM_001080509
|
TSPAN11
|
tetraspanin 11
|
|
chr17_+_43971655
|
1.886
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr1_+_95582827
|
1.877
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr4_+_154074269
|
1.873
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr11_-_123525300
|
1.850
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr12_-_48152180
|
1.843
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr5_+_140810126
|
1.832
|
NM_003735
NM_032094
|
PCDHGA12
|
protocadherin gamma subfamily A, 12
|
|
chr11_-_17410205
|
1.801
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr16_+_46919101
|
1.797
|
NM_001142466
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2
|
|
chr1_+_171154387
|
1.789
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr5_-_149669400
|
1.763
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr5_+_140855543
|
1.753
|
NM_002588
NM_032402
NM_032403
|
PCDHGC3
|
protocadherin gamma subfamily C, 3
|
|
chr4_-_100242487
|
1.749
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr5_+_63802427
|
1.744
|
NM_001029875
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein
|
|
chr5_+_140868721
|
1.695
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr2_-_217560247
|
1.695
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr20_-_41818464
|
1.688
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr5_+_140800536
|
1.670
|
NM_018914
NM_032091
NM_032092
|
PCDHGA11
|
protocadherin gamma subfamily A, 11
|
|
chr15_-_90222572
|
1.654
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr16_-_77468930
|
1.653
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr20_-_48184652
|
1.651
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr11_+_120195837
|
1.649
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chrX_+_2746810
|
1.645
|
NM_001079855
NM_001184702
NM_001184703
NM_001184704
NM_003918
|
GYG2
|
glycogenin 2
|
|
chr16_-_10674443
|
1.633
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr12_-_102872329
|
1.613
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_+_140743755
|
1.606
|
NM_018918
NM_032054
|
PCDHGA5
|
protocadherin gamma subfamily A, 5
|
|
chr17_+_47653176
|
1.598
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr1_+_161228516
|
1.588
|
NM_001102566
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
|
chr4_+_148402068
|
1.586
|
NM_001166055
NM_001957
|
EDNRA
|
endothelin receptor type A
|
|
chr5_+_149997205
|
1.579
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr10_-_99393175
|
1.523
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr7_+_45614081
|
1.515
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr9_-_91793681
|
1.506
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr11_-_17410863
|
1.503
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr5_+_140797243
|
1.496
|
NM_018927
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr7_-_11871735
|
1.480
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr8_+_30241943
|
1.474
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr2_+_241375030
|
1.473
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr11_+_69455837
|
1.466
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr10_-_90967048
|
1.449
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr20_-_50384899
|
1.446
|
NM_006045
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr5_-_125930604
|
1.445
|
NM_001182
NM_001201377
NM_001202404
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr2_-_56151273
|
1.434
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr14_+_38677186
|
1.430
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr5_+_140792507
|
1.426
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chr11_+_124789145
|
1.407
|
NM_001037558
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1
|
|
chr17_+_36584692
|
1.399
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr15_+_71433787
|
1.394
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr10_-_97321094
|
1.386
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr22_+_35937351
|
1.383
|
NM_014310
|
RASD2
|
RASD family, member 2
|
|
chr12_-_113574020
|
1.379
|
NM_001193520
NM_004658
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr6_+_151646634
|
1.376
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr5_+_140710203
|
1.375
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr4_+_154125589
|
1.363
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_+_201450730
|
1.358
|
NM_001159
|
AOX1
|
aldehyde oxidase 1
|
|
chr10_-_97200895
|
1.357
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr5_+_140767402
|
1.353
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr5_+_172068188
|
1.345
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr6_-_138893666
|
1.342
|
NM_020464
|
NHSL1
|
NHS-like 1
|
|
chr3_+_14860468
|
1.327
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr6_-_99797530
|
1.327
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr12_-_48152860
|
1.327
|
NM_001098531
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr10_-_99393851
|
1.322
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr11_+_74862031
|
1.318
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr1_-_202612580
|
1.318
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr12_-_16760935
|
1.317
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr6_+_151561094
|
1.309
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr14_+_24837225
|
1.305
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr4_-_152147520
|
1.303
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr1_-_31381436
|
1.301
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr4_+_77356252
|
1.301
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr4_-_152096655
|
1.298
|
NM_001128924
|
SH3D19
|
SH3 domain containing 19
|
|
chr1_-_68299047
|
1.294
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr11_-_76381787
|
1.293
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr10_-_118501981
|
1.261
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr13_+_113623496
|
1.256
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr5_+_140777603
|
1.254
|
NM_018925
NM_032099
|
PCDHGB5
|
protocadherin gamma subfamily B, 5
|
|
chr2_-_20424627
|
1.251
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr1_+_159141376
|
1.248
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr9_+_101705710
|
1.230
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr8_+_17396285
|
1.230
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr2_-_183731279
|
1.226
|
NM_001463
|
FRZB
|
frizzled-related protein
|
|
chr6_+_150690027
|
1.225
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr7_-_131241321
|
1.218
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_34656367
|
1.217
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr12_-_102874373
|
1.215
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_160039946
|
1.210
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr6_-_138820578
|
1.209
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr5_+_140734767
|
1.201
|
NM_018917
NM_032053
|
PCDHGA4
|
protocadherin gamma subfamily A, 4
|
|
chr16_+_66400524
|
1.200
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr16_-_75528925
|
1.199
|
NM_021615
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6
|
|
chr12_+_50497790
|
1.199
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr3_+_12045803
|
1.197
|
NM_003178
NM_133625
|
SYN2
|
synapsin II
|
|
chr19_-_460995
|
1.186
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr5_+_140787769
|
1.179
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr8_+_144295067
|
1.177
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr6_+_132129154
|
1.169
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr5_+_34656595
|
1.167
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr7_-_51384458
|
1.164
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr4_-_139163222
|
1.161
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr10_-_45474210
|
1.158
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr20_-_48099178
|
1.148
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr3_+_49591897
|
1.140
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr10_-_50323419
|
1.139
|
NM_001031746
NM_144984
|
VSTM4
|
V-set and transmembrane domain containing 4
|
|
chr6_-_105584982
|
1.138
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr10_+_128593994
|
1.136
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chr1_-_203320287
|
1.129
|
NM_002023
|
FMOD
|
fibromodulin
|
|
chr19_+_54466289
|
1.126
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr7_+_30960914
|
1.123
|
NM_001185060
NM_001185061
NM_001185062
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr3_+_61547179
|
1.115
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr1_+_61542928
|
1.114
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_-_156647088
|
1.111
|
NM_006617
|
NES
|
nestin
|
|
chr19_-_49258646
|
1.109
|
NM_000148
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group)
|
|
chr11_+_74870824
|
1.105
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr11_-_35441501
|
1.104
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chrX_-_10588458
|
1.104
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr10_-_118031542
|
1.098
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr12_+_12938540
|
1.094
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr16_+_20420854
|
1.094
|
NM_017888
|
ACSM5
|
acyl-CoA synthetase medium-chain family member 5
|
|
chr11_-_79151694
|
1.089
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr2_+_46524448
|
1.077
|
NM_001430
|
EPAS1
|
endothelial PAS domain protein 1
|
|
chr3_-_57199402
|
1.076
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr18_-_48346297
|
1.067
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr11_-_33744497
|
1.063
|
NM_001127227
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr14_+_63671101
|
1.062
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr5_+_180467224
|
1.057
|
NM_152547
|
BTNL9
|
butyrophilin-like 9
|
|
chr1_-_21995793
|
1.056
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr14_+_24838301
|
1.052
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr22_-_44708730
|
1.050
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr14_-_27066959
|
1.047
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr7_+_30951414
|
1.045
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr15_+_41851153
|
1.045
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr11_-_16430425
|
1.035
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_+_204797774
|
1.034
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr12_-_16759531
|
1.032
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_149375584
|
1.031
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr15_+_51633712
|
1.025
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr1_-_163172624
|
1.023
|
NM_001195303
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr2_+_101436491
|
1.018
|
NM_002518
|
NPAS2
|
neuronal PAS domain protein 2
|
|
chr14_+_24836144
|
1.013
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr11_-_33744272
|
1.009
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chrX_-_10544852
|
1.007
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_+_16091458
|
1.005
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr11_+_118478305
|
1.003
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr22_-_36424397
|
0.999
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr18_+_47088400
|
0.996
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr1_-_163172863
|
0.996
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr8_-_143859638
|
0.994
|
NM_023946
NM_177457
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr9_+_140033588
|
0.993
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr2_+_26915390
|
0.993
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|