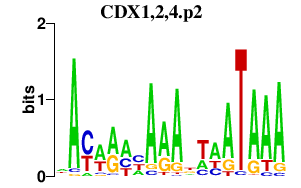
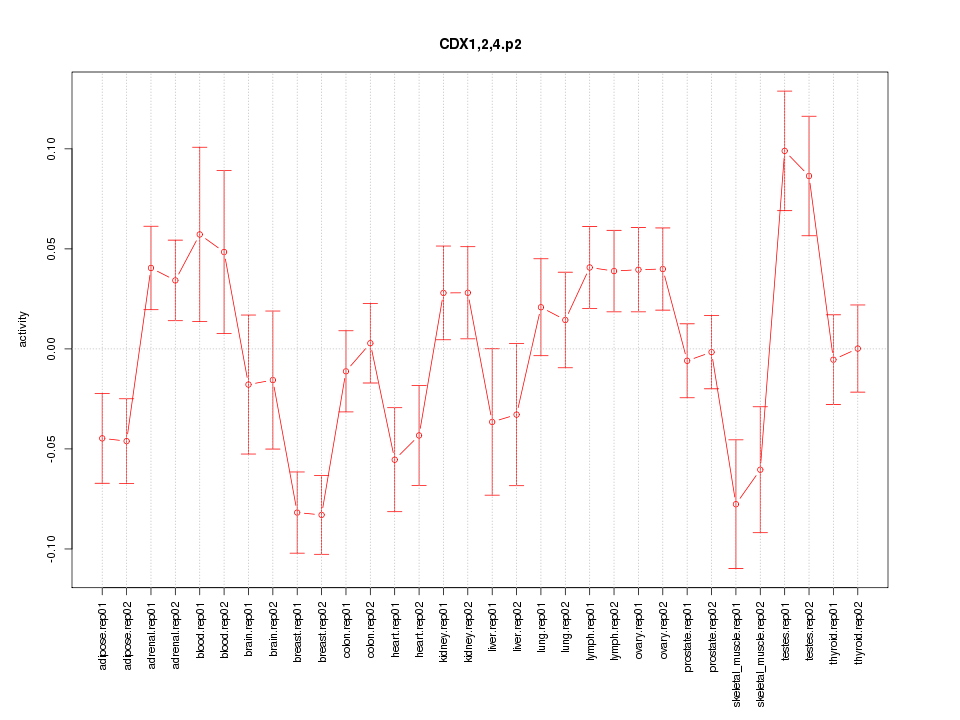
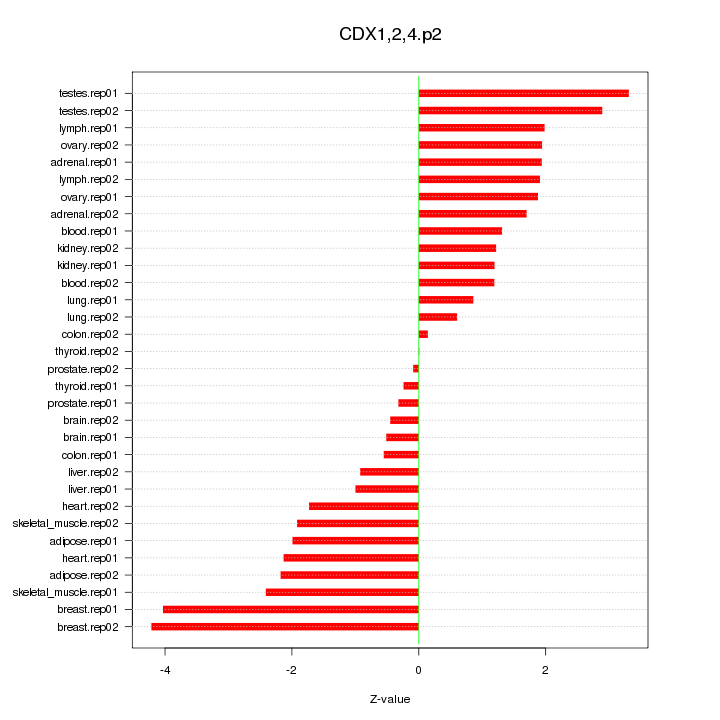
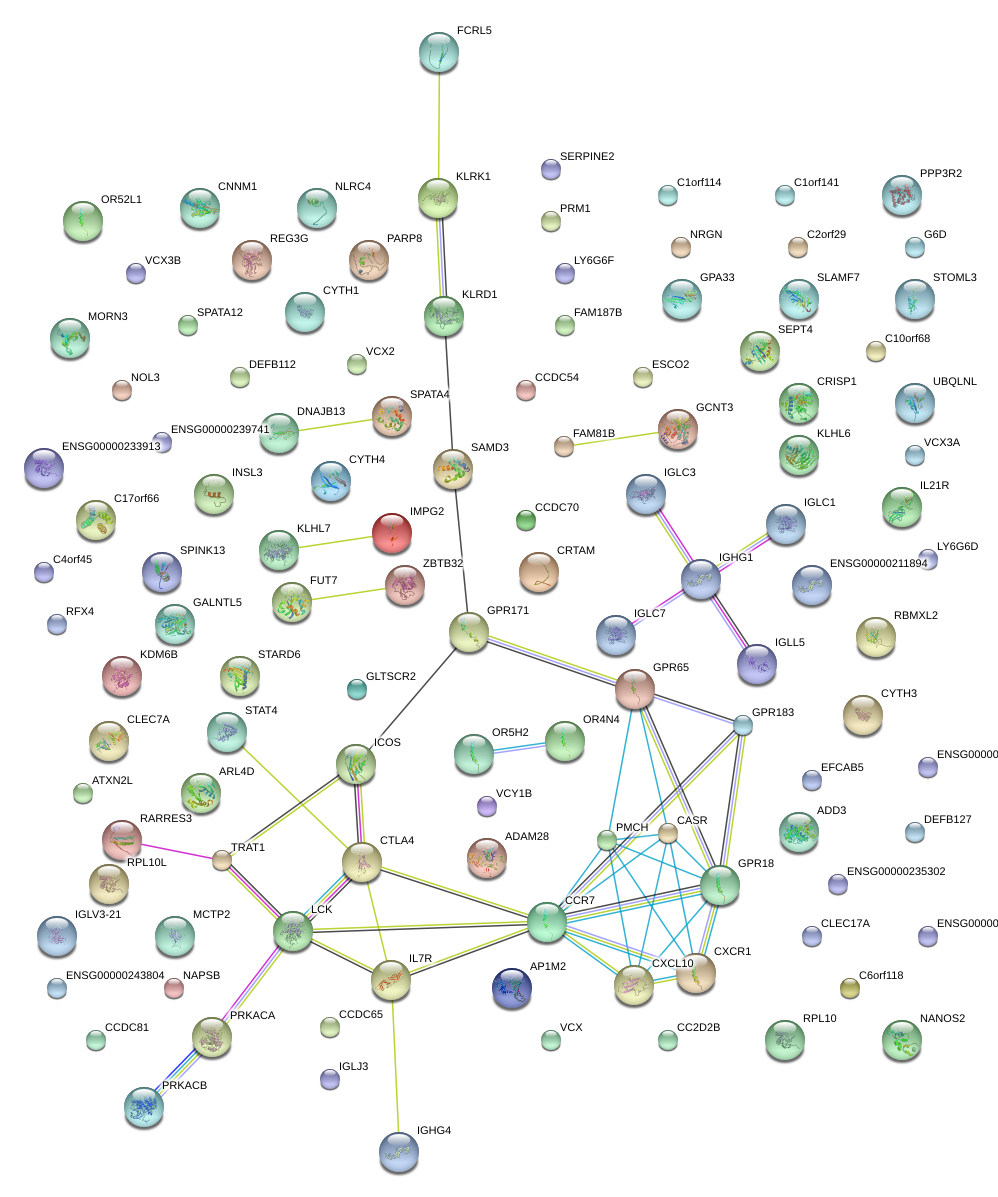
|
chr12_-_122107559
|
7.087
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr1_-_67594219
|
5.333
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr7_+_151653463
|
5.143
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr8_+_24151579
|
5.081
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr10_+_32856650
|
4.736
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr3_+_107096187
|
4.138
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr6_-_130544098
|
3.989
|
NM_001017373
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr3_+_108541544
|
3.920
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr22_+_23054862
|
3.629
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr5_+_94727047
|
3.589
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr11_+_7110164
|
3.467
|
NM_014469
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2
|
|
chr10_+_97759847
|
3.442
|
NM_001001732
NM_001159747
|
CC2D2B
|
coiled-coil and C2 domain containing 2B
|
|
chr1_+_160709038
|
3.274
|
NM_021181
|
SLAMF7
|
SLAM family member 7
|
|
chr14_-_106452693
|
3.167
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_+_57094468
|
3.104
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr6_-_49834188
|
3.047
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr17_-_34195745
|
2.992
|
NM_152781
|
C17orf66
|
chromosome 17 open reading frame 66
|
|
chr12_+_49297892
|
2.983
|
NM_033124
|
CCDC65
|
coiled-coil domain containing 65
|
|
chr9_-_139927291
|
2.877
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chr18_-_51880942
|
2.828
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr11_+_73661363
|
2.825
|
NM_153614
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr12_-_102591547
|
2.782
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr1_-_167059524
|
2.778
|
NM_005814
|
GPA33
|
glycoprotein A33 (transmembrane)
|
|
chr4_-_159956332
|
2.732
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chrX_-_6453158
|
2.649
|
NM_016379
|
VCX2
VCX3A
|
variable charge, X-linked 2
variable charge, X-linked 3A
|
|
chr12_-_10562744
|
2.647
|
NM_001199805
|
KLRC4-KLRK1
|
KLRC4-KLRK1 readthrough
|
|
chr2_-_192016293
|
2.637
|
NM_001243835
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr5_+_35856990
|
2.614
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr6_-_165723074
|
2.606
|
NM_144980
|
C6orf118
|
chromosome 6 open reading frame 118
|
|
chr5_+_147648374
|
2.590
|
NM_001040129
|
SPINK13
|
serine peptidase inhibitor, Kazal type 13 (putative)
|
|
chr19_+_36203829
|
2.581
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr3_-_183273407
|
2.552
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr8_+_27631902
|
2.540
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr11_+_122709205
|
2.511
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr16_+_27438578
|
2.499
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr3_-_101039403
|
2.448
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr4_-_76944585
|
2.440
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr4_-_177116713
|
2.371
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr19_-_50847957
|
2.369
|
|
NAPSB
|
napsin B aspartic peptidase pseudogene
|
|
chr2_-_224866616
|
2.368
|
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr12_-_10542616
|
2.365
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr14_-_47120940
|
2.336
|
NM_080746
|
RPL10L
|
ribosomal protein L10-like
|
|
chr14_+_88471467
|
2.333
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr12_-_10282680
|
2.329
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr2_+_79252825
|
2.284
|
NM_001008387
NM_198448
|
REG3G
|
regenerating islet-derived 3 gamma
|
|
chr13_-_39564857
|
2.282
|
NM_001144033
NM_145286
|
STOML3
|
stomatin (EPB72)-like 3
|
|
chr20_+_138121
|
2.271
|
NM_139074
|
DEFB127
|
defensin, beta 127
|
|
chr19_-_10697907
|
2.258
|
NM_005498
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit
|
|
chr1_-_157522151
|
2.243
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr15_+_94899163
|
2.235
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr1_+_84630575
|
2.230
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr3_-_150920946
|
2.201
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr13_-_99910548
|
2.187
|
NM_001098200
NM_005292
|
GPR18
|
G protein-coupled receptor 18
|
|
chr12_+_107078476
|
2.184
|
NM_032491
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr13_-_99959649
|
2.107
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr19_-_35719578
|
2.103
|
NM_152481
|
FAM187B
|
family with sequence similarity 187, member B
|
|
chr10_+_101088794
|
2.093
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr17_+_28268622
|
2.093
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr19_-_17932312
|
2.077
|
NM_005543
|
INSL3
|
insulin-like 3 (Leydig cell)
|
|
chr2_+_204801453
|
2.072
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr1_+_32739711
|
2.066
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr3_+_121902529
|
2.059
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr22_+_37678519
|
2.058
|
|
CYTH4
|
cytohesin 4
|
|
chr6_-_130536417
|
2.052
|
NM_152552
|
SAMD3
|
sterile alpha motif domain containing 3
|
|
chr15_+_22382381
|
1.995
|
NM_001005241
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4
|
|
chr17_-_38721701
|
1.968
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr11_-_5537815
|
1.962
|
NM_145053
|
UBQLNL
|
ubiquilin-like
|
|
chr11_+_86085777
|
1.903
|
NM_001156474
NM_021827
|
CCDC81
|
coiled-coil domain containing 81
|
|
chr13_+_52436116
|
1.884
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr11_+_63304272
|
1.883
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr22_+_37678565
|
1.868
|
|
CYTH4
|
cytohesin 4
|
|
chr12_+_10457044
|
1.866
|
NM_001114396
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr15_+_59908908
|
1.861
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr2_-_32490688
|
1.857
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr19_-_46417952
|
1.838
|
NM_001029861
|
NANOS2
|
nanos homolog 2 (Drosophila)
|
|
chr5_+_49961732
|
1.828
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr6_-_50016363
|
1.825
|
NM_001037498
|
DEFB112
|
defensin, beta 112
|
|
chr9_-_104357202
|
1.824
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr19_+_14693895
|
1.814
|
NM_001204118
NM_207390
|
CLEC17A
|
C-type lectin domain family 17, member A
|
|
chr10_+_111765725
|
1.807
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr2_-_219031646
|
1.801
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr16_-_11375094
|
1.800
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr6_+_31683084
|
1.781
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr1_-_197115566
|
1.774
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr17_-_34524054
|
1.771
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr4_-_38784588
|
1.769
|
NM_001017388
NM_001195106
NM_001195107
NM_001195108
NM_030956
|
TLR10
|
toll-like receptor 10
|
|
chr10_+_35464514
|
1.753
|
NM_182769
NM_182770
|
CREM
|
cAMP responsive element modulator
|
|
chr6_+_144185572
|
1.751
|
NM_001013623
|
C6orf94
|
chromosome 6 open reading frame 94
|
|
chr20_+_56725982
|
1.747
|
NM_178456
|
C20orf85
|
chromosome 20 open reading frame 85
|
|
chr14_-_99737821
|
1.739
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr7_+_141695678
|
1.730
|
NM_004668
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase)
|
|
chr18_+_616671
|
1.707
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr5_-_114515733
|
1.694
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr20_+_123230
|
1.688
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr2_+_204571379
|
1.685
|
|
CD28
|
CD28 molecule
|
|
chr16_+_11439279
|
1.677
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chrX_+_135730335
|
1.673
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr11_-_104905849
|
1.670
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr17_+_61086897
|
1.657
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr5_-_146460991
|
1.656
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr1_+_43613593
|
1.654
|
NM_001101376
|
FAM183A
|
family with sequence similarity 183, member A
|
|
chr12_+_54892567
|
1.649
|
NM_001184976
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr17_+_9479943
|
1.644
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chr9_-_100700422
|
1.640
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr5_+_57787260
|
1.637
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr1_+_198608136
|
1.637
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr11_-_104916041
|
1.635
|
NM_001017534
NM_052889
|
CARD16
|
caspase recruitment domain family, member 16
|
|
chr7_-_38313232
|
1.628
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr18_-_3874688
|
1.627
|
NM_001242762
NM_001242763
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr19_-_54604070
|
1.619
|
NM_130771
NM_133168
NM_133169
NM_206818
|
OSCAR
|
osteoclast associated, immunoglobulin-like receptor
|
|
chrX_-_49965003
|
1.617
|
NM_139289
|
AKAP4
|
A kinase (PRKA) anchor protein 4
|
|
chr19_+_6464259
|
1.607
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr17_+_34087915
|
1.603
|
NM_145272
|
C17orf50
|
chromosome 17 open reading frame 50
|
|
chr4_-_39033981
|
1.600
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr2_+_204732509
|
1.596
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr8_+_66933790
|
1.595
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr7_+_6793739
|
1.595
|
NM_173565
NM_001099697
|
RSPH10B
RSPH10B2
|
radial spoke head 10 homolog B (Chlamydomonas)
radial spoke head 10 homolog B2 (Chlamydomonas)
|
|
chr10_-_50342052
|
1.592
|
NM_001164484
|
FAM170B
|
family with sequence similarity 170, member B
|
|
chr17_-_5138049
|
1.584
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr1_-_24239758
|
1.583
|
NM_001841
|
CNR2
|
cannabinoid receptor 2 (macrophage)
|
|
chr12_-_53600999
|
1.583
|
NM_000889
|
ITGB7
|
integrin, beta 7
|
|
chr10_+_91498724
|
1.573
|
|
KIF20B
|
kinesin family member 20B
|
|
chr1_+_192544856
|
1.567
|
NM_002922
|
RGS1
|
regulator of G-protein signaling 1
|
|
chr22_-_17302555
|
1.565
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr19_-_14785626
|
1.556
|
NM_032571
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr2_+_90211704
|
1.551
|
|
|
|
|
chr1_+_40942878
|
1.549
|
|
ZNF642
|
zinc finger protein 642
|
|
chr1_-_190446701
|
1.547
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr15_-_55800431
|
1.544
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|
|
chr10_-_98031154
|
1.538
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr17_+_18647325
|
1.520
|
NM_031456
|
FBXW10
|
F-box and WD repeat domain containing 10
|
|
chrX_+_83116153
|
1.517
|
NM_021118
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1
|
|
chr22_+_37678423
|
1.500
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr1_-_151826042
|
1.495
|
NM_182578
|
THEM5
|
thioesterase superfamily member 5
|
|
chr11_-_102496049
|
1.486
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr5_+_40909558
|
1.468
|
NM_000587
|
C7
|
complement component 7
|
|
chr12_-_10562142
|
1.460
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 readthrough
killer cell lectin-like receptor subfamily C, member 4
|
|
chr19_-_54824343
|
1.457
|
NM_021250
NM_181879
NM_181985
NM_181986
|
LILRA5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5
|
|
chr12_-_95945226
|
1.452
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr19_-_50666279
|
1.443
|
NM_152358
|
IZUMO2
|
IZUMO family member 2
|
|
chr12_-_45307710
|
1.439
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr2_+_204571197
|
1.430
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr19_-_53794874
|
1.429
|
NM_033341
|
BIRC8
|
baculoviral IAP repeat containing 8
|
|
chrX_+_22291029
|
1.423
|
NM_152577
|
ZNF645
|
zinc finger protein 645
|
|
chr16_+_67381252
|
1.423
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chr11_+_6260321
|
1.420
|
NM_001037329
|
CNGA4
|
cyclic nucleotide gated channel alpha 4
|
|
chr1_+_198608216
|
1.418
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_36065052
|
1.404
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr11_-_17191287
|
1.402
|
NM_002645
|
PIK3C2A
|
phosphoinositide-3-kinase, class 2, alpha polypeptide
|
|
chr16_-_53405040
|
1.402
|
NM_001207030
|
LOC643802
|
u3 small nucleolar ribonucleoprotein protein MPP10-like
|
|
chr8_+_92261515
|
1.400
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr6_-_49712055
|
1.396
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chrX_-_62570968
|
1.395
|
NM_001012968
|
SPIN4
|
spindlin family, member 4
|
|
chr1_+_47137496
|
1.390
|
NM_001145474
|
ATPAF1-AS1
|
ATPAF1 antisense RNA 1 (non-protein coding)
|
|
chrX_+_23018077
|
1.383
|
NM_182699
|
DDX53
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 53
|
|
chr1_+_34326075
|
1.382
|
NM_145205
|
HMGB4
|
high mobility group box 4
|
|
chr15_+_97326629
|
1.378
|
NM_173499
|
SPATA8
|
spermatogenesis associated 8
|
|
chr12_-_53601051
|
1.374
|
|
ITGB7
|
integrin, beta 7
|
|
chr19_-_55458864
|
1.368
|
NM_001127255
NM_139176
NM_206828
|
NLRP7
|
NLR family, pyrin domain containing 7
|
|
chr20_+_68312
|
1.366
|
NM_153325
|
DEFB125
|
defensin, beta 125
|
|
chr9_-_16727887
|
1.362
|
|
BNC2
|
basonuclin 2
|
|
chr1_-_108231122
|
1.361
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr4_+_78432905
|
1.356
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr8_-_124749573
|
1.355
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chrX_+_49216616
|
1.352
|
NM_012196
NM_001472
NM_001474
NM_001475
NM_021123
NM_001477
NM_001127200
NM_001127212
|
GAGE8
GAGE1
GAGE2C
GAGE4
GAGE5
GAGE6
GAGE7
GAGE12I
GAGE2E
GAGE12B
GAGE2A
|
G antigen 8
G antigen 1
G antigen 2C
G antigen 4
G antigen 5
G antigen 6
G antigen 7
G antigen 12I
G antigen 2E
G antigen 12B
G antigen 2A
|
|
chr14_-_21029089
|
1.348
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chrX_-_44202834
|
1.344
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr7_-_104567089
|
1.342
|
|
LOC723809
|
uncharacterized LOC723809
|
|
chr9_-_88874518
|
1.336
|
NM_001010907
|
C9orf153
|
chromosome 9 open reading frame 153
|
|
chr1_+_117297056
|
1.336
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr4_+_41983712
|
1.334
|
NM_001029955
|
DCAF4L1
|
DDB1 and CUL4 associated factor 4-like 1
|
|
chr19_-_48495339
|
1.331
|
NM_001128326
|
BSPH1
|
binder of sperm protein homolog 1
|
|
chr13_-_60325073
|
1.330
|
|
|
|
|
chr2_-_99917638
|
1.315
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr5_-_35938736
|
1.312
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr3_-_189838673
|
1.311
|
|
LEPREL1
|
leprecan-like 1
|
|
chr6_-_24383519
|
1.310
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr7_+_73275488
|
1.308
|
NM_182504
|
WBSCR28
|
Williams-Beuren syndrome chromosome region 28
|
|
chr10_-_85931723
|
1.303
|
|
LOC642934
|
uncharacterized LOC642934
|
|
chr12_-_10573041
|
1.301
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr20_-_17539480
|
1.300
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr18_-_61329113
|
1.296
|
NM_006919
|
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
|
|
chr4_-_185139018
|
1.286
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr7_-_141541220
|
1.286
|
NM_001008270
NM_001171951
|
PRSS37
|
protease, serine, 37
|
|
chr12_+_47610051
|
1.281
|
NM_138371
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr8_-_87755880
|
1.270
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr8_-_30706532
|
1.266
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr7_-_84751246
|
1.266
|
NM_152754
|
SEMA3D
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D
|
|
chr13_+_78109808
|
1.264
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr16_+_27078218
|
1.260
|
NM_001145545
|
C16orf82
|
chromosome 16 open reading frame 82
|
|
chr19_-_23941571
|
1.253
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chrX_+_49226137
|
1.248
|
NM_001472
NM_001098407
|
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr7_-_112726143
|
1.247
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr4_-_84035908
|
1.247
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr5_-_110062365
|
1.244
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr10_+_15001437
|
1.244
|
NM_001080836
|
MEIG1
|
meiosis expressed gene 1 homolog (mouse)
|
|
chr15_+_94841429
|
1.240
|
NM_001159643
NM_018349
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr2_+_219221578
|
1.239
|
NM_198559
|
C2orf62
|
chromosome 2 open reading frame 62
|
|
chr15_-_80263460
|
1.235
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr13_+_108921976
|
1.235
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|