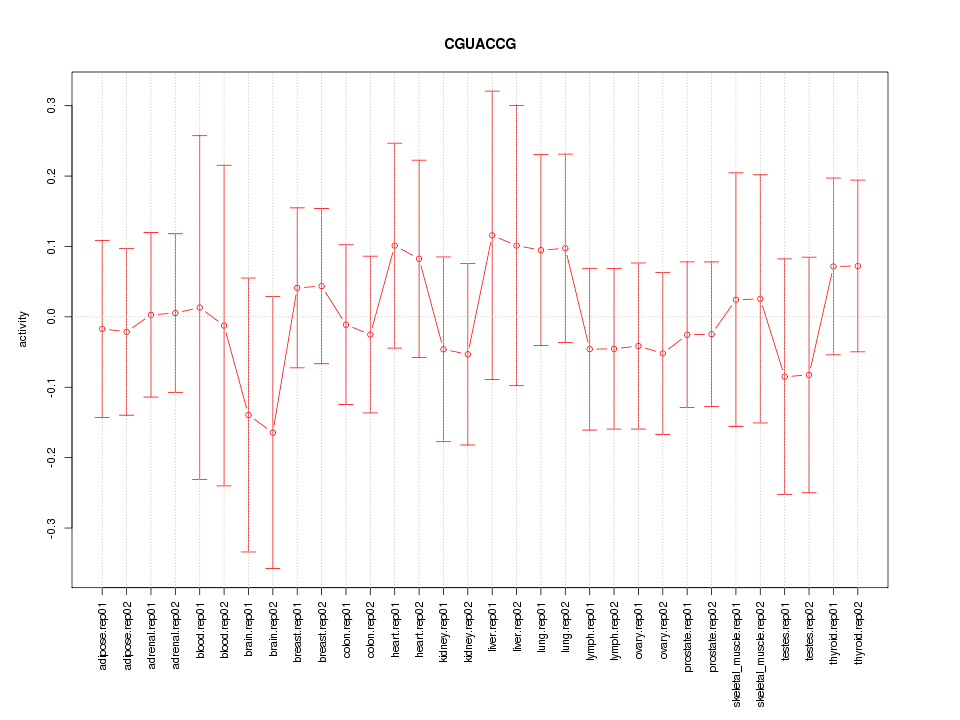
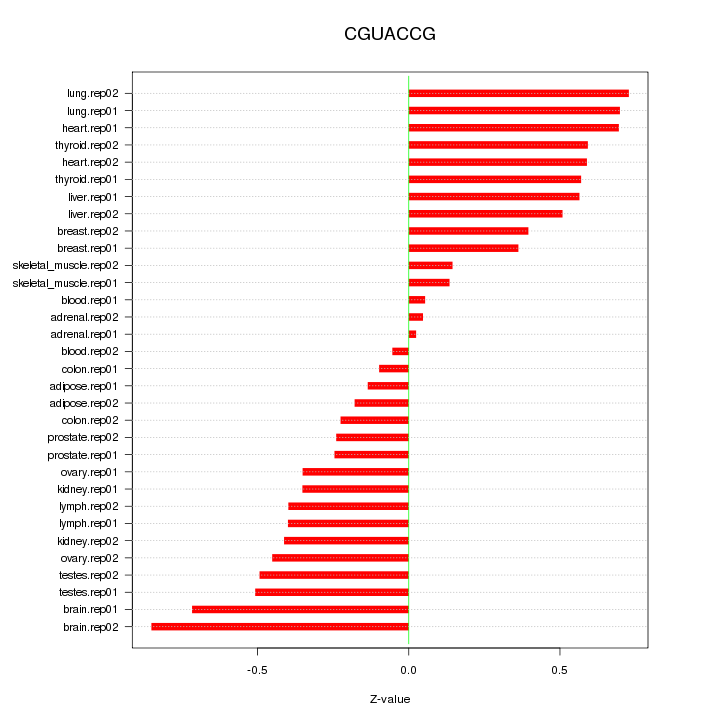
|
chr2_-_227663454
|
1.304
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr22_-_50746000
|
1.089
|
NM_012401
|
PLXNB2
|
plexin B2
|
|
chr15_+_86220267
|
0.915
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr9_+_116263706
|
0.803
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr15_+_85923739
|
0.753
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr22_+_35695267
|
0.752
|
NM_001135729
|
TOM1
|
target of myb1 (chicken)
|
|
chr14_-_21566728
|
0.736
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr6_+_108881010
|
0.735
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr14_-_21567069
|
0.652
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chr8_+_97505876
|
0.633
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr22_+_35695792
|
0.626
|
NM_001135730
NM_001135732
NM_005488
|
TOM1
|
target of myb1 (chicken)
|
|
chr9_+_139557378
|
0.620
|
NM_016215
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr14_-_21572789
|
0.608
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr9_+_139560193
|
0.577
|
NM_201446
|
EGFL7
|
EGF-like-domain, multiple 7
|
|
chr9_+_116207010
|
0.528
|
NM_144488
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr8_+_38854431
|
0.517
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr9_+_116342939
|
0.488
|
NM_021106
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr6_+_108882068
|
0.465
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr15_+_38544966
|
0.454
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr12_-_12419702
|
0.443
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr18_+_43914186
|
0.413
|
NM_152470
|
RNF165-IT1
RNF165
|
RNF165 intronic transcript 1 (non-protein coding)
ring finger protein 165
|
|
chr9_+_116327301
|
0.323
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_+_116356199
|
0.322
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr10_-_735522
|
0.293
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr5_-_57755787
|
0.240
|
NM_001252226
NM_006622
|
PLK2
|
polo-like kinase 2
|
|
chr4_-_80994309
|
0.226
|
NM_001145794
NM_058172
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr17_-_1359410
|
0.220
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr17_-_63052884
|
0.206
|
NM_006572
|
GNA13
|
guanine nucleotide binding protein (G protein), alpha 13
|
|
chr5_+_133861685
|
0.155
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr5_-_32174397
|
0.107
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr3_-_118959723
|
0.065
|
NM_003778
NM_212543
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr9_-_138798957
|
0.027
|
NM_015447
|
CAMSAP1
|
calmodulin regulated spectrin-associated protein 1
|
|
chr5_+_43121641
|
0.011
|
NM_003432
|
ZNF131
|
zinc finger protein 131
|