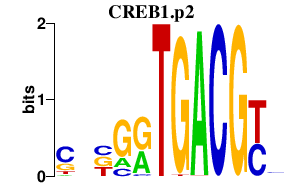
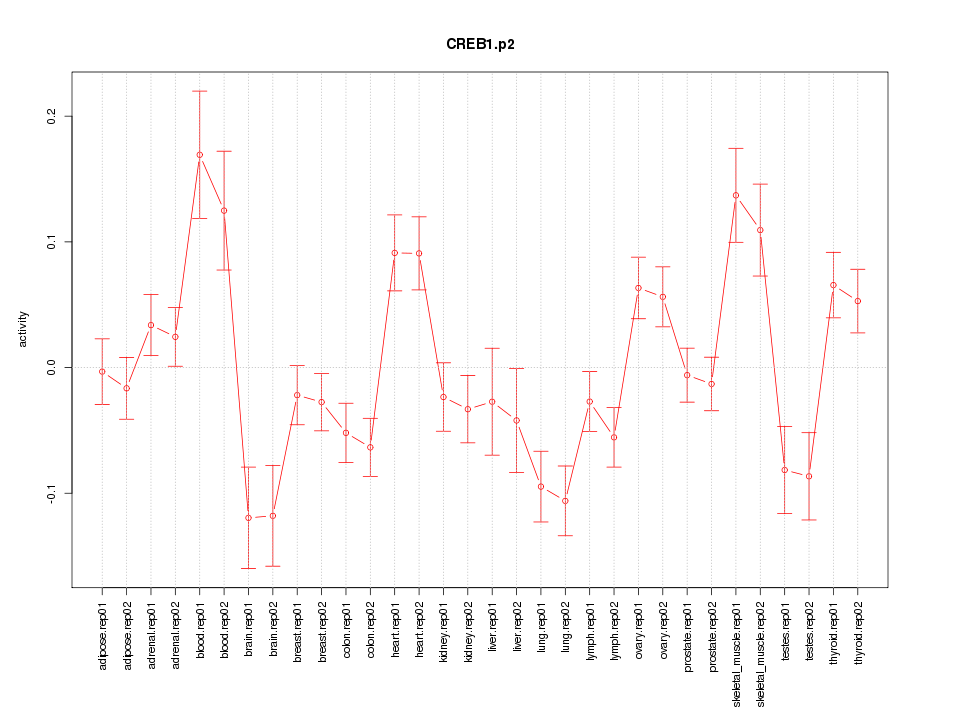
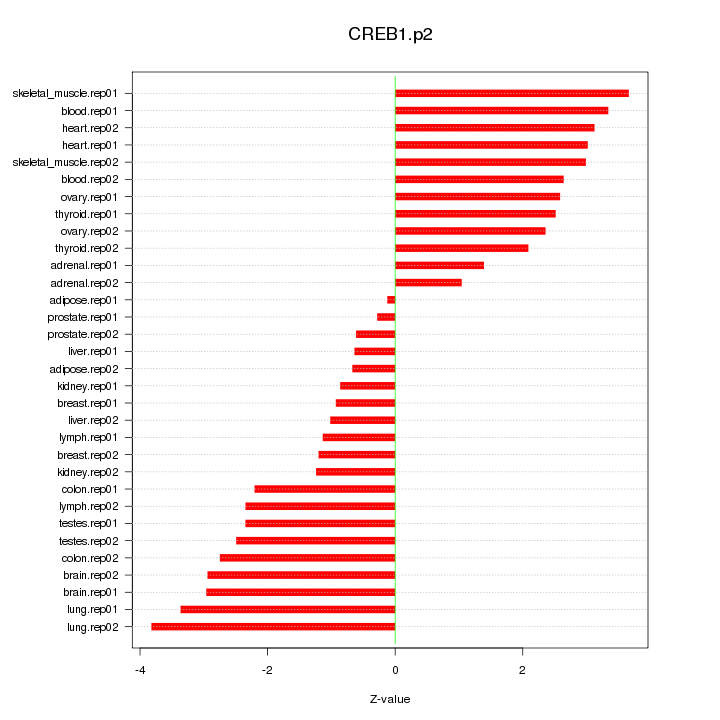
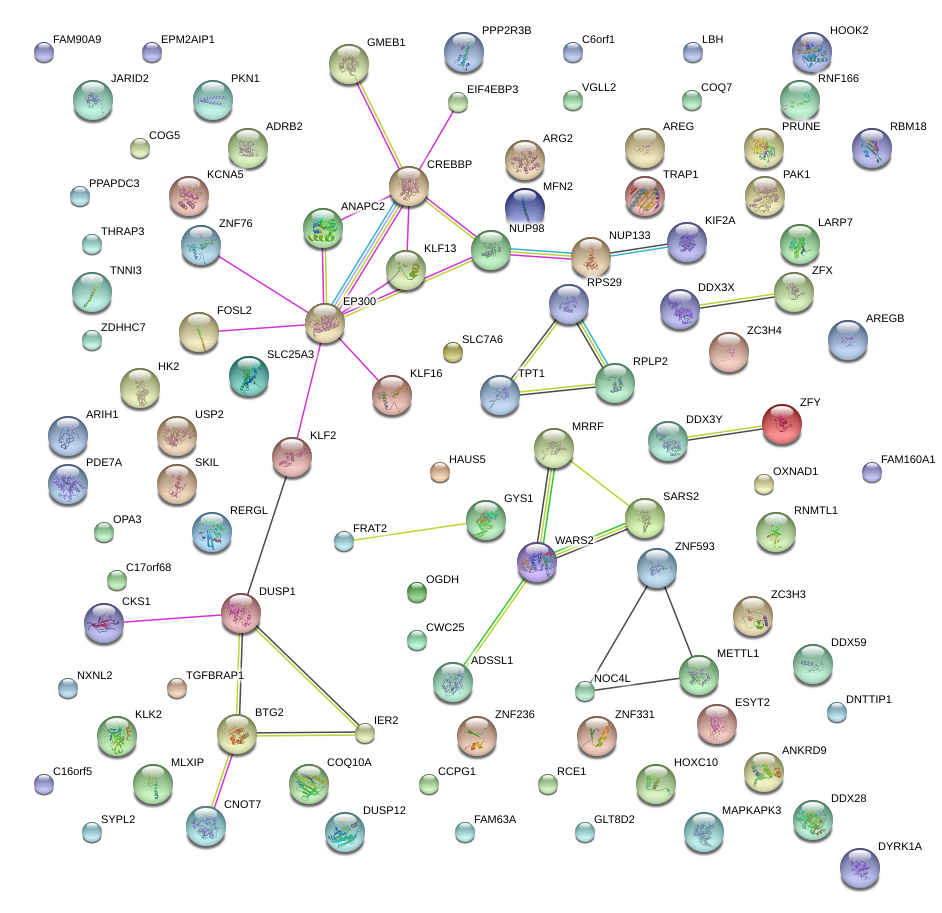
|
chr8_-_66754276
|
5.102
|
|
PDE7A
|
phosphodiesterase 7A
|
|
chr4_+_75310852
|
5.083
|
NM_001657
|
AREG
|
amphiregulin
|
|
chr6_+_15249034
|
4.755
|
|
JARID2
|
jumonji, AT rich interactive domain 2
|
|
chr19_-_46088074
|
3.799
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr3_+_50654558
|
3.624
|
NM_001243925
NM_004635
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr19_-_47617008
|
3.585
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr19_-_46088084
|
3.565
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr6_+_117586720
|
3.527
|
NM_153453
NM_182645
|
VGLL2
|
vestigial like 2 (Drosophila)
|
|
chr3_+_50654589
|
3.362
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr16_-_88772732
|
3.312
|
NM_001171815
|
RNF166
|
ring finger protein 166
|
|
chr6_+_35227234
|
3.300
|
|
ZNF76
|
zinc finger protein 76
|
|
chr16_-_3930713
|
3.282
|
|
CREBBP
|
CREB binding protein
|
|
chr6_+_35227300
|
3.199
|
|
ZNF76
|
zinc finger protein 76
|
|
chr12_+_98987389
|
3.101
|
NM_002635
NM_005888
NM_213611
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr6_+_35227478
|
3.050
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr3_+_50654868
|
3.012
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr1_+_26496382
|
2.985
|
NM_015871
|
ZNF593
|
zinc finger protein 593
|
|
chr17_-_8151353
|
2.965
|
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr1_+_12040475
|
2.944
|
|
MFN2
|
mitofusin 2
|
|
chr19_-_46088021
|
2.903
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr13_-_45915277
|
2.877
|
NM_003295
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr16_-_88772807
|
2.852
|
NM_178841
|
RNF166
|
ring finger protein 166
|
|
chr6_-_15248886
|
2.850
|
|
|
|
|
chr11_-_119252267
|
2.850
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_-_3930685
|
2.844
|
|
CREBBP
|
CREB binding protein
|
|
chr7_-_158622184
|
2.828
|
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr16_-_68057098
|
2.782
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr9_+_134165075
|
2.764
|
NM_032728
|
PPAPDC3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr13_-_45915308
|
2.736
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr16_-_68057120
|
2.716
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chrY_-_297689
|
2.655
|
NM_013239
|
PPP2R3B
|
protein phosphatase 2, regulatory subunit B'', beta
|
|
chr16_-_85045086
|
2.626
|
|
ZDHHC7
|
zinc finger, DHHC-type containing 7
|
|
chr17_+_685606
|
2.597
|
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr8_-_17104266
|
2.589
|
NM_013354
NM_054026
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr2_+_30454396
|
2.583
|
NM_030915
|
LBH
|
limb bud and heart development homolog (mouse)
|
|
chr13_-_45915203
|
2.581
|
|
TPT1
|
tumor protein, translationally-controlled 1
|
|
chr12_+_98987500
|
2.570
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chrY_+_2803409
|
2.555
|
NM_001145275
NM_003411
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr12_+_98987470
|
2.542
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr8_-_17104103
|
2.539
|
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr1_-_200638996
|
2.527
|
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr9_-_125027080
|
2.524
|
NM_033117
|
RBM18
|
RNA binding motif protein 18
|
|
chr16_+_19078916
|
2.484
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr19_-_49496564
|
2.456
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr19_-_49496369
|
2.441
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr17_-_8151373
|
2.425
|
NM_025099
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr1_-_200639091
|
2.418
|
NM_001031725
|
DDX59
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 59
|
|
chr7_-_158622202
|
2.381
|
NM_020728
|
ESYT2
|
extended synaptotagmin-like protein 2
|
|
chr12_+_98987674
|
2.371
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr1_-_119683210
|
2.366
|
NM_015836
NM_201263
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr17_+_685512
|
2.365
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr1_+_119683006
|
2.282
|
|
|
|
|
chr1_+_12040504
|
2.276
|
|
MFN2
|
mitofusin 2
|
|
chr1_+_36690038
|
2.265
|
|
THRAP3
|
thyroid hormone receptor associated protein 3
|
|
chr19_-_49496520
|
2.247
|
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr19_+_36103616
|
2.244
|
NM_015302
|
HAUS5
|
HAUS augmin-like complex, subunit 5
|
|
chr1_+_110009220
|
2.223
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr7_+_44646180
|
2.187
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr8_-_144623579
|
2.185
|
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr16_+_19078958
|
2.185
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr7_+_44646168
|
2.168
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr12_+_98987463
|
2.139
|
|
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3
|
|
chr7_+_44646197
|
2.125
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr12_+_132629014
|
2.124
|
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr9_+_125027152
|
2.099
|
|
MRRF
|
mitochondrial ribosome recycling factor
|
|
chr7_+_44646120
|
2.098
|
NM_001003941
NM_001165036
NM_002541
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr5_-_172198160
|
2.093
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr7_+_44646189
|
2.084
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr3_+_170075521
|
2.083
|
|
SKIL
|
SKI-like oncogene
|
|
chr14_+_68086633
|
2.056
|
|
ARG2
|
arginase, type II
|
|
chr9_-_125027017
|
2.056
|
|
RBM18
|
RNA binding motif protein 18
|
|
chr8_-_66753968
|
2.041
|
NM_001242318
|
PDE7A
|
phosphodiesterase 7A
|
|
chr4_+_152330326
|
2.034
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr19_+_16435734
|
2.029
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr11_-_77184549
|
2.029
|
NM_001128620
NM_002576
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr15_+_72766665
|
2.012
|
NM_005744
|
ARIH1
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila)
|
|
chr11_+_809985
|
1.997
|
|
RPLP2
|
ribosomal protein, large, P2
|
|
chr16_+_68298326
|
1.996
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr11_-_3818971
|
1.982
|
NM_005387
NM_016320
NM_139131
NM_139132
|
NUP98
|
nucleoporin 98kDa
|
|
chr12_+_122516555
|
1.970
|
NM_014938
|
MLXIP
|
MLX interacting protein
|
|
chr11_+_66610882
|
1.964
|
NM_001032279
NM_005133
|
RCE1
|
RCE1 homolog, prenyl protein protease (S. cerevisiae)
|
|
chr16_+_19079220
|
1.944
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr1_+_203274646
|
1.938
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr1_+_150980895
|
1.934
|
NM_021222
|
PRUNE
|
prune homolog (Drosophila)
|
|
chr12_+_54378929
|
1.931
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr4_+_8442496
|
1.925
|
NM_152544
|
METTL19
|
methyltransferase like 19
|
|
chr19_-_49496566
|
1.906
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chrY_+_15016775
|
1.905
|
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr14_+_105190533
|
1.895
|
NM_152328
|
ADSSL1
|
adenylosuccinate synthase like 1
|
|
chr1_-_229643996
|
1.877
|
|
NUP133
|
nucleoporin 133kDa
|
|
chr21_+_38739193
|
1.877
|
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr19_-_55668956
|
1.874
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr14_-_102975996
|
1.873
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr10_-_99094427
|
1.867
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr20_+_44420606
|
1.863
|
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr15_-_55700456
|
1.842
|
NM_004748
NM_001204450
NM_001204451
NM_020739
|
CCPG1
|
cell cycle progression 1
|
|
chrY_+_15016698
|
1.830
|
NM_004660
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr17_-_36981552
|
1.829
|
NM_017748
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr7_-_107204336
|
1.814
|
|
COG5
|
component of oligomeric golgi complex 5
|
|
chr3_+_16306666
|
1.798
|
NM_138381
|
OXNAD1
|
oxidoreductase NAD-binding domain containing 1
|
|
chr16_-_3767449
|
1.793
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr14_-_50053068
|
1.790
|
NM_001030001
NM_001032
|
RPS29
|
ribosomal protein S29
|
|
chr19_+_13263890
|
1.785
|
|
IER2
|
immediate early response 2
|
|
chr3_+_170075494
|
1.758
|
|
SKIL
|
SKI-like oncogene
|
|
chr8_-_144623611
|
1.755
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr2_+_28615668
|
1.749
|
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_+_44646220
|
1.734
|
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide)
|
|
chr16_-_4588738
|
1.729
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr12_+_56660724
|
1.704
|
|
COQ10A
|
coenzyme Q10 homolog A (S. cerevisiae)
|
|
chr9_-_140082983
|
1.699
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr3_+_170075449
|
1.689
|
NM_001145098
NM_005414
|
SKIL
|
SKI-like oncogene
|
|
chr2_+_75061234
|
1.689
|
|
HK2
|
hexokinase 2
|
|
chr5_+_139927182
|
1.687
|
NM_003732
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr4_+_113558119
|
1.685
|
NM_016648
|
LARP7
|
La ribonucleoprotein domain family, member 7
|
|
chr12_+_122516676
|
1.683
|
|
MLXIP
|
MLX interacting protein
|
|
chr19_+_54058493
|
1.683
|
NM_001079907
|
ZNF331
|
zinc finger protein 331
|
|
chr15_+_31619043
|
1.676
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr15_+_72766689
|
1.660
|
|
ARIH1
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila)
|
|
chr9_-_140083010
|
1.652
|
|
ANAPC2
|
anaphase promoting complex subunit 2
|
|
chr5_+_148206155
|
1.651
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr7_+_1200097
|
1.647
|
|
|
|
|
chr12_-_58165877
|
1.634
|
NM_005371
NM_023033
|
METTL1
|
methyltransferase like 1
|
|
chr20_+_44420610
|
1.631
|
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr8_+_128806720
|
1.629
|
|
PVT1
|
Pvt1 oncogene (non-protein coding)
|
|
chr1_-_150980840
|
1.629
|
NM_001163258
NM_018379
|
FAM63A
|
family with sequence similarity 63, member A
|
|
chr2_-_240964807
|
1.622
|
NM_004544
|
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
|
|
chr2_-_27531113
|
1.614
|
NM_003353
|
UCN
|
urocortin
|
|
chr1_-_52344010
|
1.600
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr10_+_134210691
|
1.595
|
NM_001098637
NM_138499
|
PWWP2B
|
PWWP domain containing 2B
|
|
chr3_-_52312655
|
1.591
|
NM_025222
|
WDR82
|
WD repeat domain 82
|
|
chr15_-_40401041
|
1.589
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr10_-_99094264
|
1.582
|
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr11_+_28129797
|
1.577
|
NM_001113528
NM_152636
|
METTL15
|
methyltransferase like 15
|
|
chr11_+_68228182
|
1.577
|
NM_001164160
NM_001164161
NM_001164162
NM_001164163
NM_001164164
NM_018312
|
PPP6R3
|
protein phosphatase 6, regulatory subunit 3
|
|
chr10_+_99079017
|
1.576
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr1_+_12040237
|
1.574
|
NM_001127660
NM_014874
|
MFN2
|
mitofusin 2
|
|
chr5_-_16617085
|
1.570
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr12_+_132629009
|
1.544
|
|
|
|
|
chr16_-_2014826
|
1.539
|
NM_002952
|
RPS2
|
ribosomal protein S2
|
|
chr16_+_2022081
|
1.539
|
|
TBL3
|
transducin (beta)-like 3
|
|
chr17_-_2240677
|
1.536
|
NM_018128
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr21_+_45079375
|
1.535
|
NM_015056
|
RRP1B
|
ribosomal RNA processing 1 homolog B (S. cerevisiae)
|
|
chr11_-_1785104
|
1.533
|
|
CTSD
|
cathepsin D
|
|
chr16_+_2802683
|
1.517
|
|
SRRM2
|
serine/arginine repetitive matrix 2
|
|
chr19_+_35491225
|
1.516
|
NM_001136199
NM_020895
|
GRAMD1A
|
GRAM domain containing 1A
|
|
chr17_-_685385
|
1.507
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chr3_-_71803923
|
1.504
|
NM_173359
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr22_+_41487787
|
1.503
|
|
EP300
|
E1A binding protein p300
|
|
chr7_+_120590816
|
1.499
|
NM_019071
NM_198267
|
ING3
|
inhibitor of growth family, member 3
|
|
chr22_-_47134044
|
1.497
|
|
CERK
|
ceramide kinase
|
|
chr6_-_18122850
|
1.493
|
NM_198586
|
NHLRC1
|
NHL repeat containing 1
|
|
chr3_+_50654600
|
1.491
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr2_+_61108702
|
1.486
|
|
REL
|
v-rel reticuloendotheliosis viral oncogene homolog (avian)
|
|
chr7_-_19748602
|
1.485
|
NM_001002926
|
TWISTNB
|
TWIST neighbor
|
|
chr1_-_52344306
|
1.479
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr17_+_57642885
|
1.478
|
NM_001166301
NM_024612
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_+_222791151
|
1.478
|
NM_198551
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chrX_-_46618471
|
1.477
|
NM_032591
|
SLC9A7
|
solute carrier family 9 (sodium/hydrogen exchanger), member 7
|
|
chr6_+_64281910
|
1.471
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr2_+_238536183
|
1.468
|
NM_001137550
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr11_-_1785087
|
1.461
|
|
CTSD
|
cathepsin D
|
|
chr3_+_128598520
|
1.457
|
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr15_+_40861531
|
1.455
|
NM_152260
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr3_+_44771097
|
1.452
|
NM_145044
|
ZNF501
|
zinc finger protein 501
|
|
chr11_+_117857062
|
1.451
|
NM_001558
|
IL10RA
|
interleukin 10 receptor, alpha
|
|
chr3_+_133293256
|
1.444
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr2_+_109271484
|
1.438
|
|
LIMS1
LIMS3
|
LIM and senescent cell antigen-like domains 1
LIM and senescent cell antigen-like domains 3
|
|
chr1_+_52082755
|
1.433
|
|
OSBPL9
|
oxysterol binding protein-like 9
|
|
chr6_-_26285726
|
1.430
|
NM_003543
|
HIST1H4H
|
histone cluster 1, H4h
|
|
chr19_+_42901297
|
1.428
|
|
|
|
|
chr4_-_8430185
|
1.422
|
|
ACOX3
|
acyl-CoA oxidase 3, pristanoyl
|
|
chr11_+_117049387
|
1.416
|
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr8_+_17104384
|
1.410
|
NM_001145152
NM_152415
|
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae)
|
|
chr15_+_40861527
|
1.405
|
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr3_+_38207247
|
1.403
|
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr17_-_36981405
|
1.399
|
|
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae)
|
|
chr3_+_38206968
|
1.398
|
NM_005109
|
OXSR1
|
oxidative-stress responsive 1
|
|
chr15_+_40861509
|
1.393
|
|
RPUSD2
|
RNA pseudouridylate synthase domain containing 2
|
|
chr2_-_54198055
|
1.392
|
|
|
|
|
chr1_-_229644077
|
1.390
|
NM_018230
|
NUP133
|
nucleoporin 133kDa
|
|
chr3_-_49066813
|
1.385
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chrY_+_2803111
|
1.384
|
NM_001145276
|
ZFY
|
zinc finger protein, Y-linked
|
|
chr16_+_88772884
|
1.380
|
NM_001012759
NM_001012762
|
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe)
|
|
chr17_-_20946088
|
1.376
|
NM_015276
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr11_-_64889581
|
1.373
|
NM_001997
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed
|
|
chr18_+_11851388
|
1.371
|
NM_020412
|
CHMP1B
|
charged multivesicular body protein 1B
|
|
chr14_+_105267378
|
1.365
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr17_-_76836852
|
1.364
|
NM_025090
|
USP36
|
ubiquitin specific peptidase 36
|
|
chrX_+_24167728
|
1.363
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr3_-_49066760
|
1.360
|
|
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2
|
|
chr7_+_56019552
|
1.354
|
NM_015969
|
MRPS17
|
mitochondrial ribosomal protein S17
|
|
chr12_-_76478347
|
1.353
|
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr16_-_86588680
|
1.351
|
NM_001159380
NM_001159377
NM_001159378
NM_001159379
NM_022764
|
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing
|
|
chrX_-_135055834
|
1.349
|
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr20_+_44420575
|
1.348
|
NM_052951
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr2_+_28615771
|
1.346
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr7_-_129592711
|
1.343
|
NM_001202498
|
UBE2H
|
ubiquitin-conjugating enzyme E2H
|
|
chr12_-_121790141
|
1.340
|
|
ANAPC5
|
anaphase promoting complex subunit 5
|
|
chr9_-_77643156
|
1.330
|
|
C9orf41
|
chromosome 9 open reading frame 41
|
|
chr13_-_113344050
|
1.329
|
|
LOC440149
|
uncharacterized LOC440149
|