|
chr19_+_4909447
|
10.047
|
NM_001048201
|
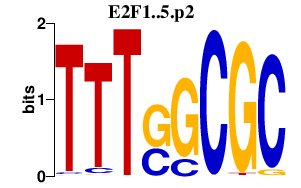
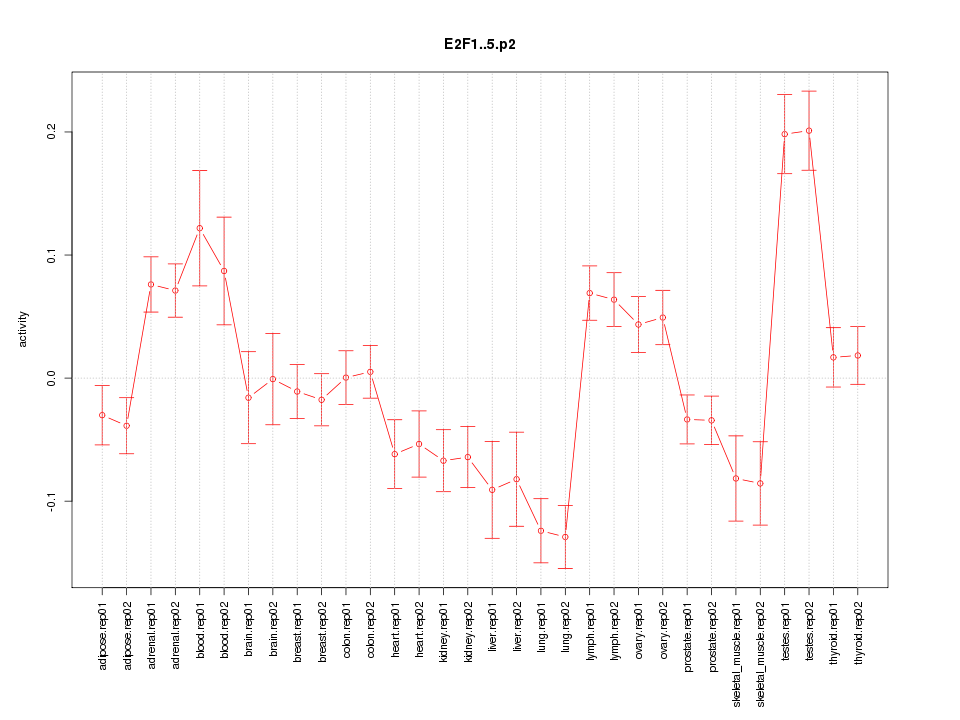
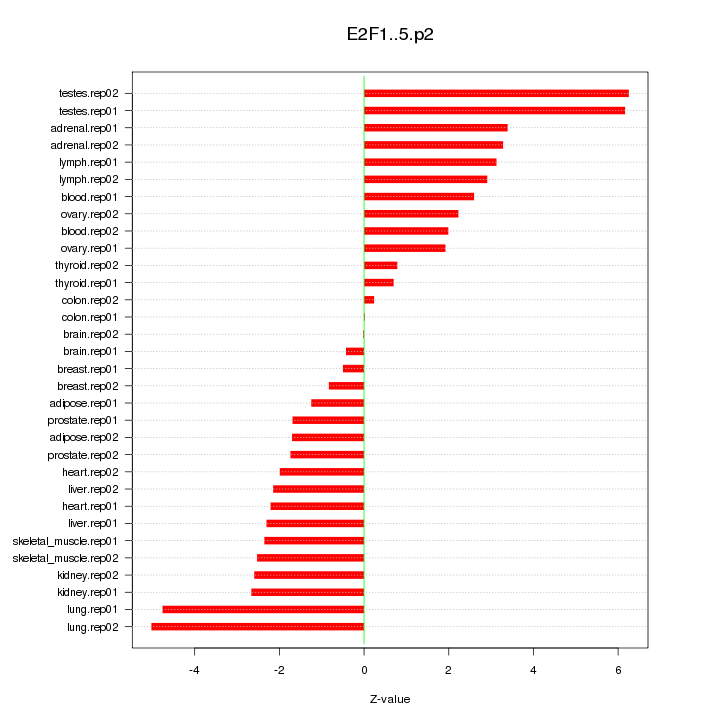
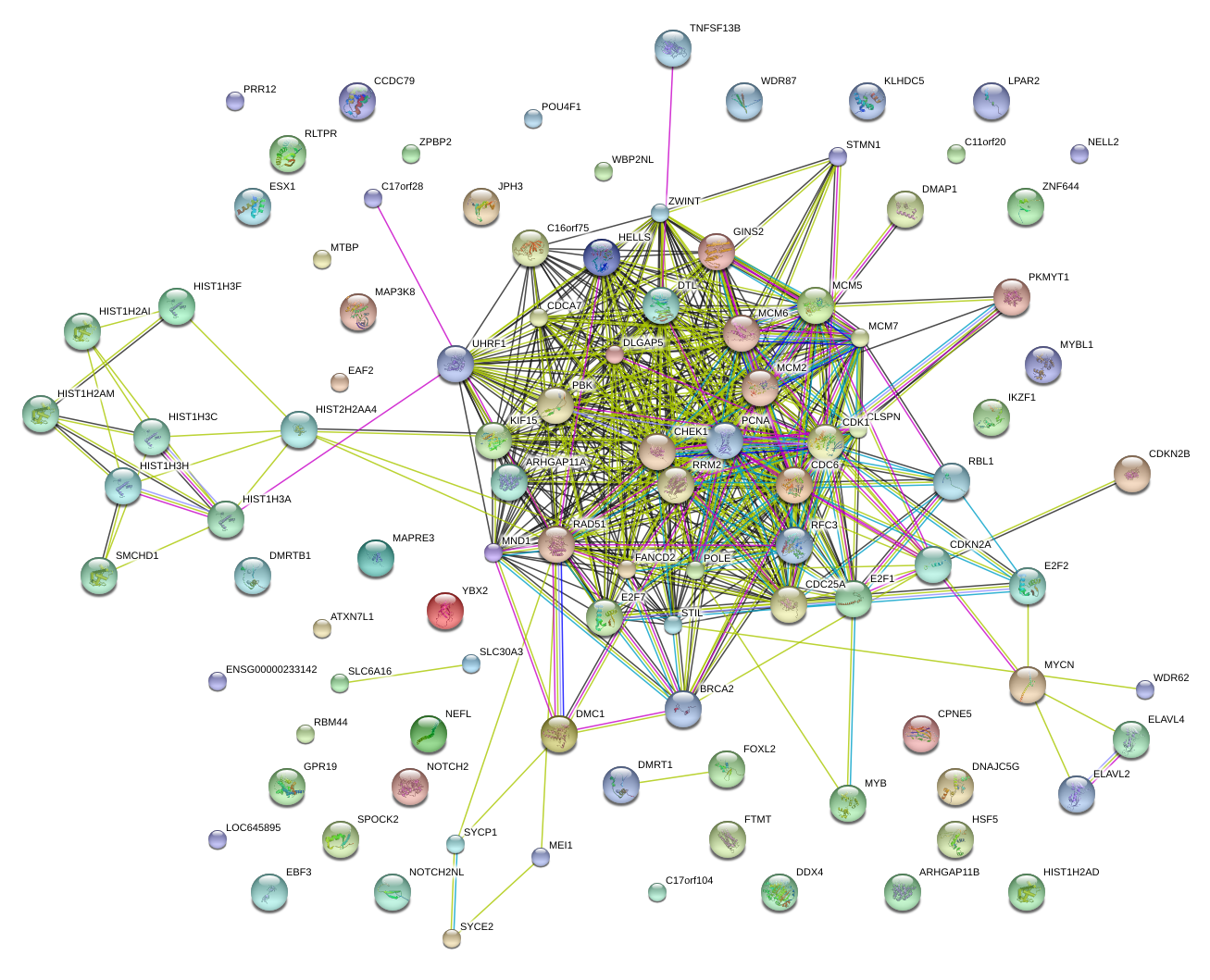
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr10_+_96305521
|
6.758
|
NM_018063
|
HELLS
|
helicase, lymphoid-specific
|
|
chr1_-_23857284
|
6.570
|
|
E2F2
|
E2F transcription factor 2
|
|
chr1_-_36235422
|
5.938
|
NM_001190481
NM_022111
|
CLSPN
|
claspin
|
|
chr1_-_23857711
|
5.811
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr2_-_27485774
|
4.373
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr6_+_135502445
|
4.336
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr6_+_135502521
|
4.218
|
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr12_-_12849120
|
4.087
|
NM_006143
|
GPR19
|
G protein-coupled receptor 19
|
|
chr20_-_32274070
|
4.070
|
|
E2F1
|
E2F transcription factor 1
|
|
chr20_-_32274154
|
3.926
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr19_+_36545757
|
3.420
|
NM_001083961
NM_173636
|
WDR62
|
WD repeat domain 62
|
|
chr10_-_131762374
|
3.158
|
|
EBF3
|
early B-cell factor 3
|
|
chr17_-_56565700
|
3.122
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr8_-_24814028
|
3.107
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr20_-_35724330
|
3.011
|
|
RBL1
|
retinoblastoma-like 1 (p107)
|
|
chr2_+_27498288
|
3.005
|
NM_173650
|
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma
|
|
chr9_+_841689
|
2.979
|
NM_021951
|
DMRT1
|
doublesex and mab-3 related transcription factor 1
|
|
chr16_+_11439279
|
2.967
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr3_+_10068097
|
2.844
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr2_-_136634002
|
2.830
|
NM_005915
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr22_+_42095502
|
2.815
|
NM_152513
|
MEI1
|
meiosis inhibitor 1
|
|
chr12_-_77459198
|
2.777
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr13_+_32889594
|
2.745
|
NM_000059
|
BRCA2
|
breast cancer 2, early onset
|
|
chr6_-_36807734
|
2.712
|
|
CPNE5
|
copine V
|
|
chr3_+_10068123
|
2.711
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr1_+_115397454
|
2.698
|
NM_003176
|
SYCP1
|
synaptonemal complex protein 1
|
|
chr8_-_67525426
|
2.695
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr2_-_136633934
|
2.689
|
|
MCM6
|
minichromosome maintenance complex component 6
|
|
chr1_+_53925482
|
2.660
|
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr10_-_58121019
|
2.622
|
NM_001005413
NM_007057
NM_032997
|
ZWINT
|
ZW10 interactor
|
|
chr3_+_121554029
|
2.577
|
NM_018456
|
EAF2
|
ELL associated factor 2
|
|
chr9_-_23821842
|
2.545
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_+_53925071
|
2.534
|
NM_033067
|
DMRTB1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr22_+_35796104
|
2.530
|
NM_006739
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr3_+_44803280
|
2.509
|
|
KIF15
|
kinesin family member 15
|
|
chr14_-_55658205
|
2.469
|
NM_001146015
NM_014750
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr19_+_4910339
|
2.437
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr1_-_26232643
|
2.436
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr8_+_121457642
|
2.430
|
NM_022045
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa
|
|
chr3_+_10068139
|
2.424
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr3_-_48229800
|
2.417
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr2_+_16080639
|
2.410
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr11_+_64067862
|
2.392
|
NM_001039496
|
C11orf20
|
chromosome 11 open reading frame 20
|
|
chr19_+_36545804
|
2.367
|
|
WDR62
|
WD repeat domain 62
|
|
chr7_+_50344255
|
2.366
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr10_+_62538088
|
2.353
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr12_-_45269340
|
2.336
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr20_-_35724401
|
2.333
|
NM_002895
NM_183404
|
RBL1
|
retinoblastoma-like 1 (p107)
|
|
chr6_+_27100784
|
2.327
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr2_+_238707387
|
2.314
|
NM_001080504
|
RBM44
|
RNA binding motif protein 44
|
|
chr16_+_11439311
|
2.314
|
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr2_+_10262853
|
2.245
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr22_+_35796154
|
2.229
|
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr16_-_66835522
|
2.216
|
NM_001136505
|
CCDC79
|
coiled-coil domain containing 79
|
|
chr10_-_131762017
|
2.204
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr16_-_85722503
|
2.157
|
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr5_+_121187649
|
2.129
|
NM_177478
|
FTMT
|
ferritin mitochondrial
|
|
chr12_-_45270071
|
2.110
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr8_-_27695288
|
2.077
|
NM_018492
|
PBK
|
PDZ binding kinase
|
|
chr1_-_26233367
|
2.069
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr11_+_125495853
|
2.055
|
|
CHEK1
|
checkpoint kinase 1
|
|
chr10_-_58121011
|
2.053
|
|
ZWINT
|
ZW10 interactor
|
|
chr10_+_62538250
|
2.045
|
|
CDK1
|
cyclin-dependent kinase 1
|
|
chrX_-_103499559
|
2.044
|
NM_153448
|
ESX1
|
ESX homeobox 1
|
|
chr2_+_174219537
|
2.040
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr19_-_49828434
|
2.039
|
NM_014037
|
SLC6A16
|
solute carrier family 6, member 16
|
|
chr1_-_47779651
|
2.035
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr17_+_38024416
|
2.015
|
NM_198844
NM_199321
|
ZPBP2
|
zona pellucida binding protein 2
|
|
chr19_-_38397233
|
1.981
|
NM_031951
|
WDR87
|
WD repeat domain 87
|
|
chr9_-_23821808
|
1.968
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_-_19738973
|
1.966
|
NM_004720
|
LPAR2
|
lysophosphatidic acid receptor 2
|
|
chr19_-_13030074
|
1.958
|
NM_001105578
|
SYCE2
|
synaptonemal complex central element protein 2
|
|
chr13_-_48877796
|
1.938
|
|
|
|
|
chr7_-_105516923
|
1.931
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr19_+_36545848
|
1.926
|
|
WDR62
|
WD repeat domain 62
|
|
chr3_+_127317212
|
1.917
|
NM_004526
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_26232890
|
1.911
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr12_-_45270157
|
1.903
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr16_+_87636492
|
1.891
|
NM_020655
|
JPH3
|
junctophilin 3
|
|
chr16_+_67679011
|
1.889
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr13_-_79177682
|
1.869
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr3_+_127317274
|
1.864
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr6_+_27775898
|
1.862
|
NM_003509
|
HIST1H2AI
HIST1H2AM
HIST1H3F
|
histone cluster 1, H2ai
histone cluster 1, H2am
histone cluster 1, H3f
|
|
chr17_+_42733744
|
1.840
|
NM_001145080
|
C17orf104
|
chromosome 17 open reading frame 104
|
|
chr22_+_35796294
|
1.826
|
|
MCM5
|
minichromosome maintenance complex component 5
|
|
chr17_+_38444134
|
1.816
|
NM_001254
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr12_+_27933162
|
1.800
|
NM_020782
|
KLHDC5
|
kelch domain containing 5
|
|
chr15_+_40987326
|
1.798
|
NM_001164270
NM_002875
NM_133487
NM_001164269
|
RAD51
|
RAD51 homolog (S. cerevisiae)
|
|
chr15_+_32907690
|
1.795
|
NM_014783
NM_199357
|
ARHGAP11A
|
Rho GTPase activating protein 11A
|
|
chr1_+_212208918
|
1.788
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr1_-_120612175
|
1.767
|
|
NOTCH2
|
notch 2
|
|
chr20_-_5100443
|
1.764
|
NM_182649
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr16_+_67679058
|
1.744
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr17_-_7197866
|
1.743
|
NM_015982
|
YBX2
|
Y box binding protein 2
|
|
chr22_-_38966066
|
1.734
|
NM_007068
|
DMC1
|
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast)
|
|
chr1_-_91487543
|
1.716
|
|
ZNF644
|
zinc finger protein 644
|
|
chr10_-_73848309
|
1.711
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr9_-_21994414
|
1.691
|
NM_058195
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)
|
|
chr16_-_3030391
|
1.687
|
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr13_+_34392254
|
1.686
|
|
RFC3
|
replication factor C (activator 1) 3, 38kDa
|
|
chr10_-_73848278
|
1.684
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr22_+_42394728
|
1.677
|
NM_152613
|
WBP2NL
|
WBP2 N-terminal like
|
|
chr1_+_212209129
|
1.674
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr3_+_10028579
|
1.670
|
|
LOC442075
|
uncharacterized LOC442075
|
|
chr6_+_26045611
|
1.655
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr4_+_154265774
|
1.654
|
NM_001253861
NM_032117
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae)
|
|
chr17_-_7197811
|
1.639
|
|
YBX2
|
Y box binding protein 2
|
|
chr19_+_50094911
|
1.637
|
NM_020719
|
PRR12
|
proline rich 12
|
|
chr12_-_133263988
|
1.636
|
|
POLE
|
polymerase (DNA directed), epsilon
|
|
chr5_+_55033844
|
1.627
|
NM_001142549
NM_001166533
NM_024415
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr3_-_138665981
|
1.623
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr19_-_38878663
|
1.603
|
NM_152657
|
GGN
|
gametogenetin
|
|
chr2_+_47168362
|
1.590
|
|
TTC7A
|
tetratricopeptide repeat domain 7A
|
|
chr2_+_10262694
|
1.574
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr12_-_45269953
|
1.570
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr16_-_3030473
|
1.569
|
NM_004203
NM_182687
|
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1
|
|
chr15_+_91260575
|
1.564
|
NM_000057
|
BLM
|
Bloom syndrome, RecQ helicase-like
|
|
chr3_+_127317271
|
1.554
|
|
MCM2
|
minichromosome maintenance complex component 2
|
|
chr1_-_91487586
|
1.550
|
|
ZNF644
|
zinc finger protein 644
|
|
chr2_-_24346143
|
1.540
|
NM_199346
|
PFN4
|
profilin family, member 4
|
|
chr3_-_148804122
|
1.535
|
NM_003071
NM_139048
|
HLTF
|
helicase-like transcription factor
|
|
chr9_-_123639462
|
1.533
|
|
PHF19
|
PHD finger protein 19
|
|
chrX_+_131157231
|
1.525
|
NM_001042453
NM_016542
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr15_-_64673569
|
1.524
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr17_+_38444151
|
1.524
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr7_-_148581403
|
1.523
|
NM_001203247
NM_001203248
NM_004456
NM_152998
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr2_+_17935124
|
1.521
|
NM_182625
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr19_+_46144904
|
1.519
|
NM_001242348
|
LOC100287177
|
uncharacterized LOC100287177
|
|
chr1_+_23037329
|
1.512
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr3_+_51428720
|
1.504
|
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr11_-_64851449
|
1.492
|
|
CDCA5
|
cell division cycle associated 5
|
|
chr3_-_48229328
|
1.488
|
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr3_+_51428698
|
1.474
|
NM_013286
|
RBM15B
|
RNA binding motif protein 15B
|
|
chr3_+_44803208
|
1.474
|
NM_020242
|
KIF15
|
kinesin family member 15
|
|
chr6_+_27114860
|
1.468
|
NM_080596
|
HIST1H2AH
|
histone cluster 1, H2ah
|
|
chr6_-_27100528
|
1.464
|
NM_021058
|
HIST1H2BJ
|
histone cluster 1, H2bj
|
|
chr5_+_93954352
|
1.462
|
NM_032290
|
ANKRD32
|
ankyrin repeat domain 32
|
|
chr1_+_36038970
|
1.458
|
NM_178548
|
TFAP2E
|
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon)
|
|
chr9_-_104500861
|
1.456
|
NM_133445
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr2_-_70995113
|
1.452
|
NM_001185055
NM_001617
NM_017482
NM_017488
|
ADD2
|
adducin 2 (beta)
|
|
chr1_-_120612260
|
1.452
|
|
NOTCH2
|
notch 2
|
|
chr1_-_25256364
|
1.450
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_+_48867650
|
1.448
|
NM_012451
|
SYNGR4
|
synaptogyrin 4
|
|
chr2_+_17935379
|
1.442
|
NM_001130009
|
GEN1
|
Gen endonuclease homolog 1 (Drosophila)
|
|
chr17_+_29158948
|
1.438
|
NM_024857
|
ATAD5
|
ATPase family, AAA domain containing 5
|
|
chr4_+_178231013
|
1.434
|
|
NEIL3
|
nei endonuclease VIII-like 3 (E. coli)
|
|
chr16_-_85722587
|
1.433
|
NM_016095
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr20_+_25388320
|
1.423
|
NM_021067
|
GINS1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr1_-_120612301
|
1.418
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr19_-_6502224
|
1.415
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr7_-_100026284
|
1.405
|
NM_017984
|
ZCWPW1
|
zinc finger, CW type with PWWP domain 1
|
|
chr6_+_31707724
|
1.402
|
NM_002441
NM_025259
NM_172165
NM_172166
|
MSH5-C6orf26
MSH5
|
MSH5-C6orf26 readthrough (non-protein coding)
mutS homolog 5 (E. coli)
|
|
chr6_+_79787548
|
1.398
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr4_-_130014405
|
1.396
|
NM_144643
|
SCLT1
|
sodium channel and clathrin linker 1
|
|
chr2_+_74781511
|
1.392
|
NM_001381
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1)
|
|
chr17_+_38444130
|
1.392
|
|
CDC6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr1_-_200992824
|
1.387
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr18_+_71983109
|
1.387
|
NM_001174123
|
C18orf63
|
chromosome 18 open reading frame 63
|
|
chr17_-_33288475
|
1.383
|
NM_001193529
NM_001193530
NM_006584
|
CCT6B
|
chaperonin containing TCP1, subunit 6B (zeta 2)
|
|
chr9_-_123639566
|
1.382
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr9_+_114287438
|
1.379
|
NM_001007169
NM_133464
|
ZNF483
|
zinc finger protein 483
|
|
chr7_-_148581297
|
1.378
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr8_+_24771276
|
1.372
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_+_61542790
|
1.364
|
|
|
|
|
chr17_-_56769375
|
1.364
|
NM_001201457
NM_031272
NM_198393
|
TEX14
|
testis expressed 14
|
|
chr8_-_132052834
|
1.355
|
NM_001115
|
ADCY8
|
adenylate cyclase 8 (brain)
|
|
chr15_-_83952194
|
1.349
|
|
BNC1
|
basonuclin 1
|
|
chrX_+_131157635
|
1.342
|
NM_001042452
|
MST4
|
serine/threonine protein kinase MST4
|
|
chr6_-_27806116
|
1.331
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr13_+_34392205
|
1.330
|
NM_002915
NM_181558
|
RFC3
|
replication factor C (activator 1) 3, 38kDa
|
|
chr17_-_7197896
|
1.325
|
|
YBX2
|
Y box binding protein 2
|
|
chr20_+_25388328
|
1.318
|
|
GINS1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chrX_-_139587224
|
1.313
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr8_+_99439249
|
1.308
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr19_-_14016908
|
1.299
|
NM_024323
|
C19orf57
|
chromosome 19 open reading frame 57
|
|
chr19_-_1863546
|
1.297
|
NM_031918
|
KLF16
|
Kruppel-like factor 16
|
|
chr1_-_182922002
|
1.295
|
|
SHCBP1L
|
SHC SH2-domain binding protein 1-like
|
|
chr15_+_92397027
|
1.285
|
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1
|
|
chr7_+_94285676
|
1.281
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chrX_+_105066531
|
1.272
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr7_+_94285620
|
1.270
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr7_-_148581377
|
1.265
|
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr15_+_40987387
|
1.260
|
|
RAD51
|
RAD51 homolog (S. cerevisiae)
|
|
chr5_+_133861685
|
1.259
|
NM_015288
|
PHF15
|
PHD finger protein 15
|
|
chr10_+_112327411
|
1.258
|
NM_005445
|
SMC3
|
structural maintenance of chromosomes 3
|
|
chr6_-_36164973
|
1.258
|
|
|
|
|
chr7_+_94285736
|
1.257
|
|
PEG10
|
paternally expressed 10
|
|
chr19_+_2096792
|
1.256
|
NM_001031735
NM_001039846
|
IZUMO4
|
IZUMO family member 4
|
|
chr5_+_68485367
|
1.256
|
NM_022909
|
CENPH
|
centromere protein H
|
|
chr1_+_2518071
|
1.254
|
NM_001195736
NM_001195737
NM_001195738
NM_001195740
NM_001195741
NM_152371
|
FAM213B
|
family with sequence similarity 213, member B
|
|
chr1_-_91487769
|
1.249
|
NM_032186
NM_201269
|
ZNF644
|
zinc finger protein 644
|
|
chr1_+_151043079
|
1.249
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr1_+_38158189
|
1.247
|
|
CDCA8
|
cell division cycle associated 8
|
|
chr20_-_44563573
|
1.244
|
|
FLJ40606
|
uncharacterized LOC643549
|
|
chr1_+_240255171
|
1.243
|
NM_020066
|
FMN2
|
formin 2
|
|
chr16_+_88870214
|
1.243
|
|
CDT1
|
chromatin licensing and DNA replication factor 1
|
|
chr8_+_48873533
|
1.240
|
|
MCM4
|
minichromosome maintenance complex component 4
|
|
chr1_-_228645517
|
1.227
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr3_+_10857867
|
1.225
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|