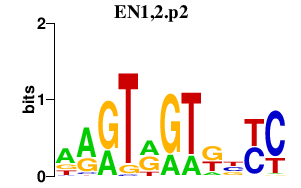
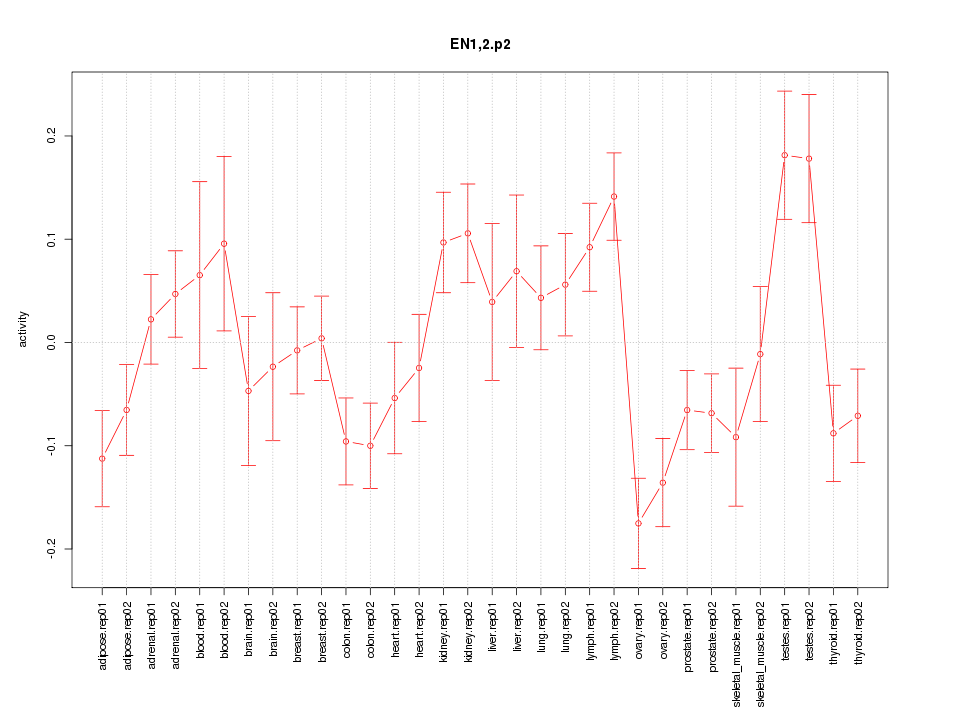
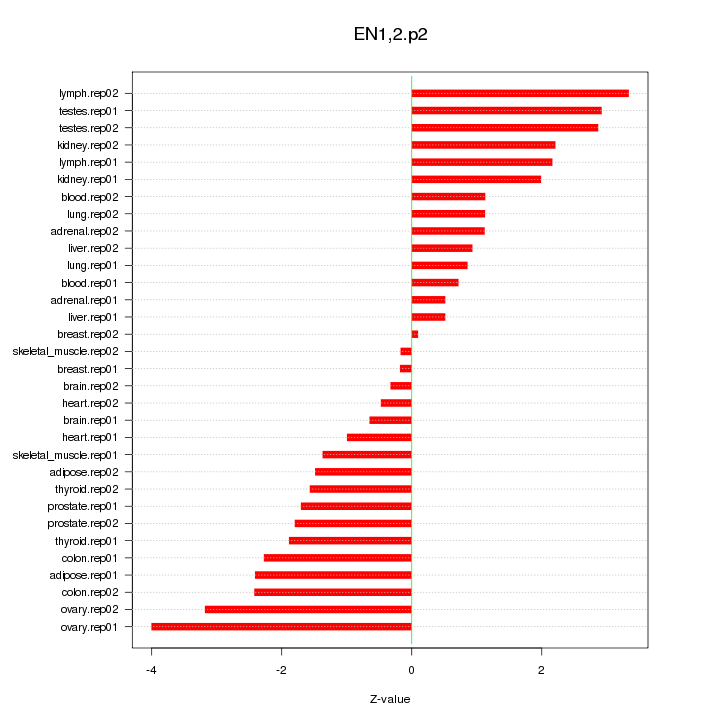
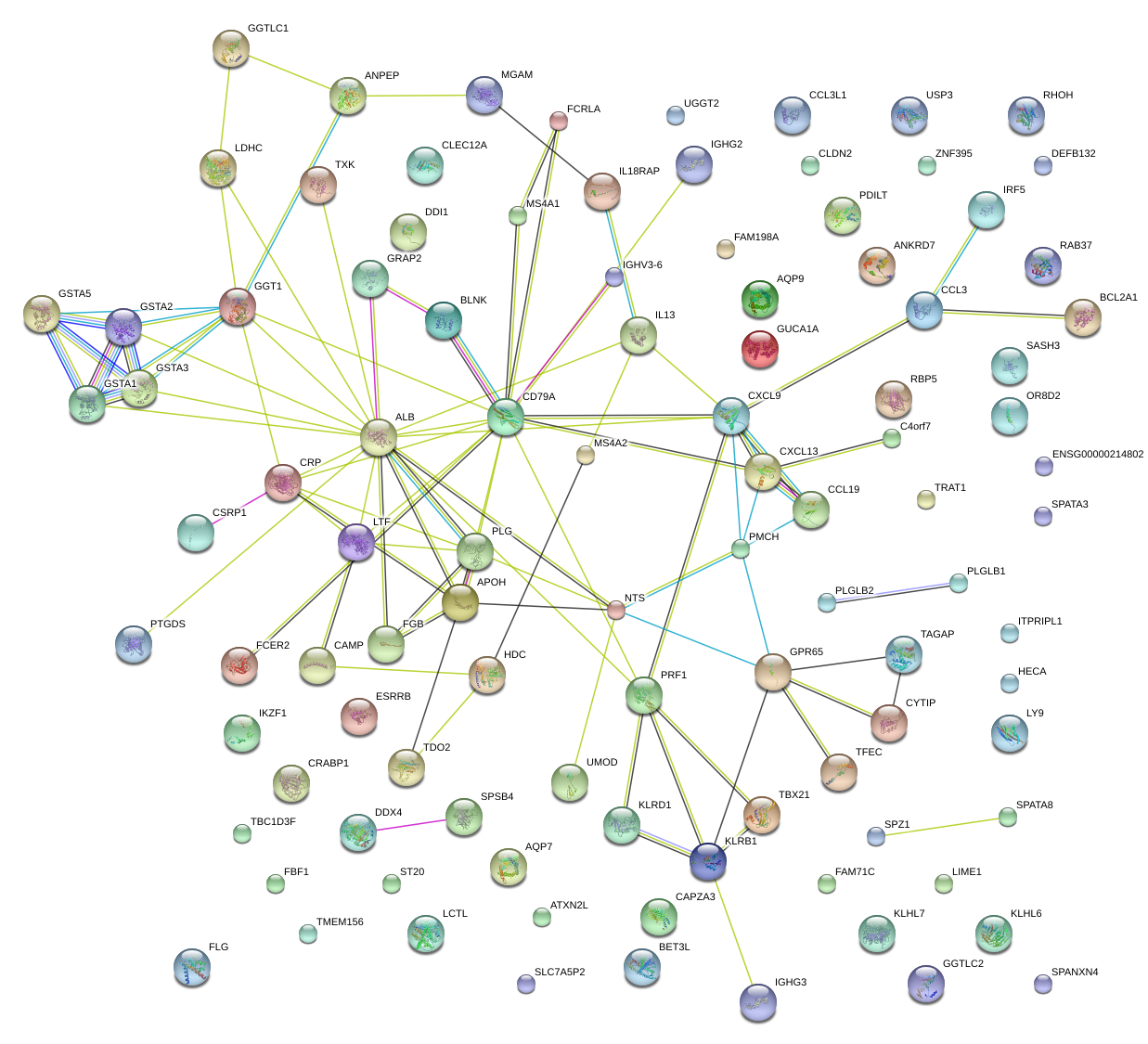
|
chr4_+_74281884
|
4.418
|
|
ALB
|
albumin
|
|
chr11_+_60223281
|
4.388
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr4_+_71200670
|
4.344
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr16_-_20364029
|
4.181
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr7_+_141695678
|
3.794
|
NM_004668
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase)
|
|
chr4_+_78432905
|
3.782
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr17_+_45810609
|
3.677
|
NM_013351
|
TBX21
|
T-box 21
|
|
chr7_+_117864711
|
3.589
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr4_+_155484131
|
3.563
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr4_+_156824846
|
3.548
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr4_+_71091787
|
3.213
|
NM_152997
|
C4orf7
|
chromosome 4 open reading frame 7
|
|
chr7_+_50348255
|
3.176
|
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr4_-_48136245
|
3.135
|
NM_003328
|
TXK
|
TXK tyrosine kinase
|
|
chr12_+_100041527
|
3.124
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr12_+_10460416
|
3.064
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr12_+_18891040
|
3.056
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr12_-_7281374
|
2.900
|
NM_031491
|
RBP5
|
retinol binding protein 5, cellular
|
|
chr2_+_88047603
|
2.883
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr2_-_87248942
|
2.852
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr12_-_102591547
|
2.770
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr4_-_39033981
|
2.604
|
NM_024943
|
TMEM156
|
transmembrane protein 156
|
|
chr14_+_88471467
|
2.604
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr1_-_159684274
|
2.576
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr3_+_108541544
|
2.409
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr10_-_98031154
|
2.355
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_96991934
|
2.333
|
NM_178495
NM_001163524
|
ITPRIPL1
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 1
|
|
chr2_+_231860838
|
2.272
|
NM_139073
|
SPATA3
|
spermatogenesis associated 3
|
|
chr4_+_40198526
|
2.212
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr22_+_40297085
|
2.205
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr20_-_23967431
|
2.197
|
NM_178312
|
GGTLC1
|
gamma-glutamyltransferase light chain 1
|
|
chr9_+_139872014
|
2.195
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr9_+_139871954
|
2.193
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr17_-_64225496
|
2.178
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr1_+_160765927
|
2.176
|
NM_001033667
NM_002348
|
LY9
|
lymphocyte antigen 9
|
|
chr12_+_86268072
|
2.166
|
NM_006183
|
NTS
|
neurotensin
|
|
chr9_-_34729463
|
2.144
|
NM_001141917
|
FAM205A
|
family with sequence similarity 205, member A
|
|
chr4_+_74269971
|
2.124
|
NM_000477
|
ALB
|
albumin
|
|
chr7_-_115670828
|
2.105
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr12_-_9760481
|
2.057
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr15_+_97326629
|
2.050
|
NM_173499
|
SPATA8
|
spermatogenesis associated 8
|
|
chr7_-_115670774
|
1.980
|
|
TFEC
|
transcription factor EC
|
|
chr3_-_183273407
|
1.952
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chrX_+_142113703
|
1.949
|
NM_001009613
|
SPANXN4
|
SPANX family, member N4
|
|
chr2_+_103035148
|
1.943
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr1_+_161676761
|
1.919
|
NM_001184866
NM_001184867
NM_001184870
NM_001184871
NM_001184872
NM_001184873
NM_032738
|
FCRLA
|
Fc receptor-like A
|
|
chr7_+_128578246
|
1.874
|
NM_032643
|
IRF5
|
interferon regulatory factor 5
|
|
chrX_+_106171328
|
1.835
|
|
CLDN2
|
claudin 2
|
|
chr6_+_42123143
|
1.828
|
NM_000409
|
GUCA1A
|
guanylate cyclase activator 1A (retina)
|
|
chrX_+_128913897
|
1.783
|
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr19_-_7764283
|
1.772
|
NM_001207019
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr11_+_103907307
|
1.772
|
NM_001001711
|
DDI1
|
DNA-damage inducible 1 homolog 1 (S. cerevisiae)
|
|
chr12_+_10163225
|
1.750
|
NM_001129998
NM_205852
|
CLEC12B
|
C-type lectin domain family 12, member B
|
|
chr4_-_76928601
|
1.703
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr6_-_52668599
|
1.693
|
NM_145740
|
GSTA1
|
glutathione S-transferase alpha 1
|
|
chr3_+_48264836
|
1.675
|
NM_004345
|
CAMP
|
cathelicidin antimicrobial peptide
|
|
chr15_-_80263460
|
1.664
|
NM_001114735
NM_004049
|
BCL2A1
|
BCL2-related protein A1
|
|
chr3_-_46505077
|
1.660
|
NM_001199149
|
LTF
|
lactotransferrin
|
|
chr17_+_72732958
|
1.655
|
NM_001163989
|
RAB37
|
RAB37, member RAS oncogene family
|
|
chr6_-_159466019
|
1.620
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr5_+_55062712
|
1.605
|
NM_001166534
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chrX_+_65235279
|
1.603
|
|
|
|
|
chr15_-_90349819
|
1.594
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr5_+_79615789
|
1.571
|
NM_032567
|
SPZ1
|
spermatogenic leucine zipper 1
|
|
chr20_+_62367945
|
1.568
|
NM_017806
|
LIME1
|
Lck interacting transmembrane adaptor 1
|
|
chr15_-_50558161
|
1.542
|
NM_002112
|
HDC
|
histidine decarboxylase
|
|
chr19_+_42381189
|
1.532
|
NM_001783
NM_021601
|
CD79A
|
CD79a molecule, immunoglobulin-associated alpha
|
|
chr15_+_58430394
|
1.525
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chrX_+_128913881
|
1.489
|
NM_018990
|
SASH3
|
SAM and SH3 domain containing 3
|
|
chr6_-_116866772
|
1.489
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr11_+_18433845
|
1.486
|
NM_002301
NM_017448
|
LDHC
|
lactate dehydrogenase C
|
|
chr5_+_131993864
|
1.481
|
NM_002188
|
IL13
|
interleukin 13
|
|
chr17_-_34417386
|
1.476
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr22_+_40297183
|
1.476
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr2_-_158300517
|
1.460
|
NM_004288
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr10_-_72362513
|
1.451
|
NM_001083116
NM_005041
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr14_+_76837689
|
1.435
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr16_-_20416032
|
1.426
|
NM_174924
|
PDILT
|
protein disulfide isomerase-like, testis expressed
|
|
chr15_-_66857704
|
1.421
|
NM_207338
|
LCTL
|
lactase-like
|
|
chr3_+_186714335
|
1.414
|
|
|
|
|
chr9_-_34691135
|
1.409
|
NM_006274
|
CCL19
|
chemokine (C-C motif) ligand 19
|
|
chr3_+_140770742
|
1.406
|
NM_080862
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr20_+_238376
|
1.397
|
NM_207469
|
DEFB132
|
defensin, beta 132
|
|
chr3_-_120401401
|
1.355
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr11_+_124134699
|
1.352
|
NM_001005198
|
OR8G5
|
olfactory receptor, family 8, subfamily G, member 5
|
|
chr16_-_28518131
|
1.349
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr12_-_49999259
|
1.348
|
NM_032130
|
FAM186B
|
family with sequence similarity 186, member B
|
|
chr2_+_182850550
|
1.343
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr12_+_56732658
|
1.340
|
NM_016584
|
IL23A
|
interleukin 23, alpha subunit p19
|
|
chr19_-_42348507
|
1.339
|
NM_173506
|
LYPD4
|
LY6/PLAUR domain containing 4
|
|
chr3_+_186435097
|
1.339
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chrX_-_104465349
|
1.332
|
NM_031274
|
TEX13A
|
testis expressed 13A
|
|
chr1_+_6105980
|
1.331
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr17_-_38956146
|
1.328
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr5_+_57787260
|
1.302
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr6_+_31674639
|
1.287
|
NM_001003693
|
LY6G6F
|
lymphocyte antigen 6 complex, locus G6F
|
|
chr6_+_31583766
|
1.287
|
NM_032955
|
AIF1
|
allograft inflammatory factor 1
|
|
chr11_-_62752370
|
1.277
|
NM_004790
NM_153276
NM_153277
NM_153278
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6
|
|
chr2_-_192015706
|
1.272
|
|
STAT4
|
signal transducer and activator of transcription 4
|
|
chr1_+_212208918
|
1.267
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr2_+_135596185
|
1.260
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr6_-_25726754
|
1.256
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr5_-_66492574
|
1.253
|
NM_005582
|
CD180
|
CD180 molecule
|
|
chr1_+_192605267
|
1.250
|
NM_002927
NM_144766
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr12_+_6554032
|
1.246
|
NM_001242
|
CD27
|
CD27 molecule
|
|
chr1_-_157789832
|
1.232
|
NM_001159397
NM_001159398
NM_052938
|
FCRL1
|
Fc receptor-like 1
|
|
chr1_+_57320442
|
1.230
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr17_-_4643162
|
1.224
|
NM_001100812
NM_022059
|
CXCL16
|
chemokine (C-X-C motif) ligand 16
|
|
chr5_-_176836535
|
1.221
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr22_-_37545944
|
1.218
|
NM_000878
|
IL2RB
|
interleukin 2 receptor, beta
|
|
chr1_-_161039413
|
1.213
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr6_+_25727004
|
1.211
|
NM_170610
|
HIST1H2BA
|
histone cluster 1, H2ba
|
|
chr6_+_116782555
|
1.197
|
NM_001010919
|
FAM26F
|
family with sequence similarity 26, member F
|
|
chr6_-_41006927
|
1.197
|
NM_173561
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like
|
|
chr12_-_18890917
|
1.180
|
NM_033123
|
PLCZ1
|
phospholipase C, zeta 1
|
|
chr11_-_124180661
|
1.174
|
NM_001002917
|
OR8D1
|
olfactory receptor, family 8, subfamily D, member 1
|
|
chr10_-_72362441
|
1.164
|
|
PRF1
|
perforin 1 (pore forming protein)
|
|
chr13_+_52436116
|
1.161
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chrX_+_49593905
|
1.160
|
NM_007003
|
PAGE4
|
P antigen family, member 4 (prostate associated)
|
|
chr2_+_204801453
|
1.158
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chrX_-_47489703
|
1.152
|
NM_002621
|
CFP
|
complement factor properdin
|
|
chr14_-_23588819
|
1.141
|
NM_001805
|
CEBPE
|
CCAAT/enhancer binding protein (C/EBP), epsilon
|
|
chr12_-_95945226
|
1.139
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr19_-_14169970
|
1.135
|
NM_001145028
|
PALM3
|
paralemmin 3
|
|
chr19_-_4302381
|
1.126
|
NM_001169126
NM_144615
|
TMIGD2
|
transmembrane and immunoglobulin domain containing 2
|
|
chr1_+_212209129
|
1.115
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr6_+_161123224
|
1.115
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr3_-_120400957
|
1.113
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr1_+_159259503
|
1.112
|
NM_002001
|
FCER1A
|
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide
|
|
chr6_-_87804773
|
1.111
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr5_+_175108463
|
1.107
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr1_+_34326075
|
1.093
|
NM_145205
|
HMGB4
|
high mobility group box 4
|
|
chr12_+_113344811
|
1.081
|
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa
|
|
chr1_-_152086555
|
1.080
|
NM_007113
|
TCHH
|
trichohyalin
|
|
chr2_+_234668914
|
1.078
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr18_+_72201799
|
1.072
|
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family)
|
|
chr7_-_38389575
|
1.062
|
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr16_+_67381252
|
1.052
|
NM_001161575
|
LRRC36
|
leucine rich repeat containing 36
|
|
chrX_-_127186339
|
1.051
|
NM_138289
|
ACTRT1
|
actin-related protein T1
|
|
chr2_+_79347610
|
1.050
|
|
REG1A
|
regenerating islet-derived 1 alpha
|
|
chr9_-_104145800
|
1.047
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr12_-_55378455
|
1.047
|
NM_001136030
|
KIAA0748
|
KIAA0748
|
|
chr9_+_105757592
|
1.043
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr11_-_47399986
|
1.042
|
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene spi1
|
|
chr1_-_108231122
|
1.037
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr11_-_26743545
|
1.033
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr15_-_34629044
|
1.031
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr17_+_43922255
|
1.028
|
NM_175882
|
IMP5
|
intramembrane protease 5
|
|
chr1_+_212209112
|
1.028
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr5_-_135701163
|
1.011
|
NM_001167576
NM_001167577
NM_020389
|
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7
|
|
chr6_-_52628272
|
1.000
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr20_+_34556528
|
0.999
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr1_+_16348365
|
0.999
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr11_+_118764100
|
0.997
|
NM_032966
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr17_-_47286742
|
0.995
|
NM_031498
NM_001198755
NM_001198756
|
GNGT2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2
|
|
chr11_+_122709205
|
0.989
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr12_-_10542616
|
0.972
|
NM_007360
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1
|
|
chr6_+_14117857
|
0.968
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr2_+_88824168
|
0.968
|
NM_152670
|
C2orf51
|
chromosome 2 open reading frame 51
|
|
chr20_-_44175995
|
0.961
|
NM_001198986
NM_020398
|
SPINLW1-WFDC6
SPINLW1
|
SPINLW1-WFDC6 readthrough
serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin)
|
|
chr7_-_115670811
|
0.960
|
|
TFEC
|
transcription factor EC
|
|
chr10_+_45406629
|
0.956
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chrX_+_107288199
|
0.954
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr3_+_118892363
|
0.953
|
NM_006952
|
UPK1B
|
uroplakin 1B
|
|
chr19_+_10959105
|
0.951
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr1_+_212797788
|
0.947
|
NM_153606
|
FAM71A
|
family with sequence similarity 71, member A
|
|
chr12_-_123201319
|
0.945
|
NM_006018
|
HCAR3
|
hydroxycarboxylic acid receptor 3
|
|
chr1_-_205391180
|
0.944
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr22_-_37215516
|
0.940
|
NM_002854
|
PVALB
|
parvalbumin
|
|
chr1_-_27701302
|
0.939
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chr19_-_54327428
|
0.936
|
NM_144687
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr11_+_118215058
|
0.917
|
NM_000073
|
CD3G
|
CD3g molecule, gamma (CD3-TCR complex)
|
|
chr12_-_9885859
|
0.911
|
NM_001253748
NM_001253750
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr8_-_87755880
|
0.910
|
NM_019098
|
CNGB3
|
cyclic nucleotide gated channel beta 3
|
|
chr8_+_32785485
|
0.907
|
|
|
|
|
chr6_-_133084585
|
0.903
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr3_+_52813936
|
0.901
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr1_+_244214552
|
0.900
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr4_+_186350543
|
0.888
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr6_-_51952368
|
0.883
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr4_+_159442865
|
0.883
|
NM_001253727
NM_001253728
NM_001253729
NM_001253730
NM_001253732
NM_001253733
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr11_+_5968576
|
0.882
|
NM_001003443
|
OR56A3
|
olfactory receptor, family 56, subfamily A, member 3
|
|
chr18_-_58040000
|
0.881
|
NM_005912
|
MC4R
|
melanocortin 4 receptor
|
|
chr2_+_87144737
|
0.880
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr5_-_176326314
|
0.878
|
NM_002115
|
HK3
|
hexokinase 3 (white cell)
|
|
chr1_-_76397975
|
0.876
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chrY_+_16168097
|
0.874
|
NM_181880
NM_004679
|
VCY1B
VCX3A
VCY
|
variable charge, Y-linked 1B
variable charge, X-linked 3A
variable charge, Y-linked
|
|
chr22_+_23054862
|
0.874
|
|
IGLV3-21
IGLJ3
IGLC1
|
immunoglobulin lambda variable 3-21
immunoglobulin lambda joining 3
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr14_+_95027760
|
0.871
|
NM_006215
|
SERPINA4
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4
|
|
chr22_+_50986461
|
0.865
|
NM_138433
|
KLHDC7B
|
kelch domain containing 7B
|
|
chr10_-_48416692
|
0.862
|
NM_016204
|
GDF2
|
growth differentiation factor 2
|
|
chr4_-_74847669
|
0.857
|
NM_002619
|
PF4
|
platelet factor 4
|
|
chr1_+_111742946
|
0.853
|
|
|
|
|
chr16_+_57050985
|
0.851
|
NM_032206
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr1_-_161601752
|
0.850
|
NM_000570
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr12_+_8608521
|
0.847
|
NM_001007033
|
CLEC6A
|
C-type lectin domain family 6, member A
|
|
chrX_+_49226137
|
0.841
|
NM_001472
NM_001098407
|
GAGE2C
GAGE2E
GAGE2D
GAGE2A
|
G antigen 2C
G antigen 2E
G antigen 2D
G antigen 2A
|
|
chr12_-_11548485
|
0.841
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr4_-_74853819
|
0.833
|
NM_002704
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7)
|
|
chr11_-_45928832
|
0.828
|
NM_001080446
|
C11orf94
|
chromosome 11 open reading frame 94
|
|
chr1_-_161039703
|
0.825
|
|
ARHGAP30
|
Rho GTPase activating protein 30
|