|
chr10_-_92680784
|
4.754
|
|
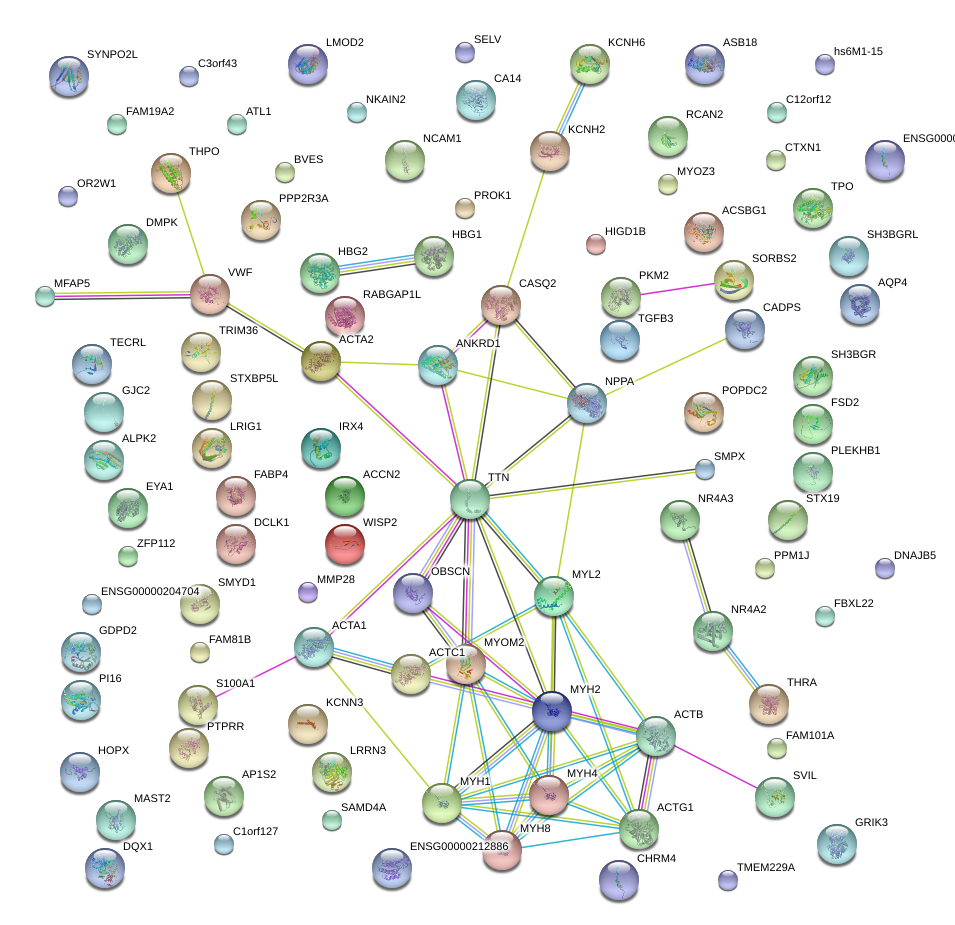
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr7_-_150652861
|
3.852
|
NM_001204798
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr12_-_111358353
|
3.634
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr8_+_1993157
|
3.493
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr1_-_11907741
|
3.404
|
NM_006172
|
NPPA
|
natriuretic peptide A
|
|
chr1_+_228395740
|
3.296
|
NM_001098623
NM_052843
|
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr4_-_65275000
|
3.131
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr10_-_92681025
|
2.853
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr1_-_116311161
|
2.661
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr5_+_150040402
|
2.464
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr15_-_35087748
|
2.313
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr3_-_62861012
|
2.304
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr21_+_40823779
|
2.227
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr8_-_72268720
|
2.125
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_+_110731061
|
2.092
|
NM_001099658
NM_001099660
NM_018334
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr20_+_43343523
|
2.045
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr7_-_123673486
|
2.045
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr11_+_73358593
|
2.024
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr2_-_74753311
|
1.996
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr10_-_29754478
|
1.950
|
|
SVIL
|
supervillin
|
|
chr19_+_32083186
|
1.944
|
NM_001205273
|
THEG5
|
testis highly expressed protein 5
|
|
chr12_+_124773709
|
1.911
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr15_+_63889551
|
1.884
|
NM_203373
|
FBXL22
|
F-box and leucine-rich repeat protein 22
|
|
chr1_-_37499708
|
1.840
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr15_-_83474805
|
1.816
|
NM_001007122
|
FSD2
|
fibronectin type III and SPRY domain containing 2
|
|
chr5_+_94727047
|
1.797
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr2_+_88367298
|
1.788
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr9_+_102584136
|
1.786
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr1_+_46379258
|
1.724
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chrX_+_69642880
|
1.673
|
NM_001171191
NM_001171192
NM_001171193
NM_017711
|
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2
|
|
chr19_-_44831907
|
1.625
|
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr7_+_123295860
|
1.588
|
NM_207163
|
LMOD2
|
leiomodin 2 (cardiac)
|
|
chr6_+_124125068
|
1.583
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr1_-_113257873
|
1.548
|
NM_005167
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J
|
|
chr1_-_11042093
|
1.538
|
NM_001170754
|
C1orf127
|
chromosome 1 open reading frame 127
|
|
chr19_-_46285752
|
1.517
|
NM_001081560
NM_001081562
NM_004409
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr12_-_71148383
|
1.501
|
NM_130846
NM_001207016
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr11_+_112831968
|
1.499
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr3_-_93747453
|
1.497
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr4_-_186732208
|
1.480
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_120627026
|
1.469
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr18_-_56296188
|
1.462
|
NM_052947
|
ALPK2
|
alpha-kinase 2
|
|
chr1_-_154832187
|
1.458
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr9_+_34990715
|
1.425
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chrX_-_21776158
|
1.410
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr3_-_66551341
|
1.393
|
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr2_-_179672056
|
1.392
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr12_-_91348950
|
1.379
|
NM_152638
|
C12orf12
|
chromosome 12 open reading frame 12
|
|
chr1_+_153600859
|
1.371
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr12_+_50451486
|
1.369
|
NM_001095
NM_020039
|
ACCN2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr10_-_75410778
|
1.367
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr6_+_36922208
|
1.336
|
NM_153370
|
PI16
|
peptidase inhibitor 16
|
|
chr17_-_10452939
|
1.285
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chrX_-_15873094
|
1.272
|
NM_003916
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr1_+_228337414
|
1.268
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr2_-_237172987
|
1.267
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr14_+_55034633
|
1.266
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr12_-_71148063
|
1.261
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr19_-_46285746
|
1.247
|
|
DMPK
|
dystrophia myotonica-protein kinase
|
|
chr6_-_105584542
|
1.245
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chrX_-_15872938
|
1.236
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr3_-_196242190
|
1.207
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr2_+_1418179
|
1.182
|
NM_175721
NM_175722
|
TPO
|
thyroid peroxidase
|
|
chr18_-_24442534
|
1.179
|
NM_004028
|
AQP4
|
aquaporin 4
|
|
chr4_-_186732047
|
1.174
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_174843523
|
1.159
|
NM_001243763
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr11_-_46408105
|
1.159
|
NM_000741
|
CHRM4
|
cholinergic receptor, muscarinic 4
|
|
chrX_-_15872905
|
1.140
|
|
|
|
|
chr1_+_110993748
|
1.123
|
NM_032414
|
PROK1
|
prokineticin 1
|
|
chr2_-_157189040
|
1.097
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr5_-_1882764
|
1.096
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chrX_-_15872921
|
1.073
|
|
AP1S2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr3_+_135741576
|
1.048
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr3_-_119379269
|
1.010
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr6_-_46459710
|
0.994
|
NM_001251973
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_-_7990974
|
0.982
|
NM_206833
|
CTXN1
|
cortexin 1
|
|
chr5_+_88185090
|
0.980
|
|
|
|
|
chr9_+_34990266
|
0.974
|
NM_001135005
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr14_-_76447235
|
0.957
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr17_-_34122547
|
0.956
|
NM_001032278
NM_024302
|
MMP28
|
matrix metallopeptidase 28
|
|
chr5_-_114515733
|
0.948
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr11_-_5271035
|
0.947
|
NM_000559
|
HBG1
|
hemoglobin, gamma A
|
|
chr6_-_29012951
|
0.944
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chr17_+_42925278
|
0.940
|
NM_016438
|
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B
|
|
chr1_+_150230168
|
0.925
|
NM_012113
|
CA14
|
carbonic anhydrase XIV
|
|
chr15_-_72501017
|
0.914
|
|
PKM2
|
pyruvate kinase, muscle
|
|
chr12_-_8815370
|
0.901
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr14_+_51026742
|
0.897
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr13_-_36429798
|
0.883
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_-_57547845
|
0.881
|
NM_001145460
NM_032495
NM_139212
|
HOPX
|
HOP homeobox
|
|
chr17_+_38230678
|
0.872
|
|
THRA
|
thyroid hormone receptor, alpha
|
|
chr8_-_82395401
|
0.872
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr12_-_6233684
|
0.869
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr12_-_62586547
|
0.846
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr17_+_38024416
|
0.831
|
NM_198844
NM_199321
|
ZPBP2
|
zona pellucida binding protein 2
|
|
chr14_+_55034329
|
0.830
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr3_+_10857867
|
0.822
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr12_+_26348488
|
0.817
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr11_-_93583667
|
0.814
|
NM_001144871
|
VSTM5
|
V-set and transmembrane domain containing 5
|
|
chr6_-_36807217
|
0.811
|
NM_020939
|
CPNE5
|
copine V
|
|
chr7_+_97361270
|
0.803
|
NM_003182
NM_013996
NM_013997
NM_013998
|
TAC1
|
tachykinin, precursor 1
|
|
chr12_+_131438451
|
0.795
|
NM_198827
|
GPR133
|
G protein-coupled receptor 133
|
|
chr6_-_26108320
|
0.789
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr11_-_74442429
|
0.787
|
|
CHRDL2
|
chordin-like 2
|
|
chr12_-_7848324
|
0.780
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr21_+_25801042
|
0.778
|
|
FLJ42200
|
FLJ42200 protein
|
|
chr11_-_5275904
|
0.776
|
NM_000184
|
HBG1
HBG2
|
hemoglobin, gamma A
hemoglobin, gamma G
|
|
chr19_-_44405934
|
0.773
|
|
LOC100505715
|
uncharacterized LOC100505715
|
|
chr11_-_74442185
|
0.769
|
NM_015424
|
CHRDL2
|
chordin-like 2
|
|
chr6_-_46293215
|
0.769
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr22_-_38479148
|
0.765
|
NM_013356
|
SLC16A8
|
solute carrier family 16, member 8 (monocarboxylic acid transporter 3)
|
|
chr4_-_177116713
|
0.757
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr2_-_30144431
|
0.756
|
NM_004304
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase
|
|
chr5_+_113698304
|
0.750
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr11_-_62689011
|
0.748
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr19_-_46417952
|
0.748
|
NM_001029861
|
NANOS2
|
nanos homolog 2 (Drosophila)
|
|
chr20_+_17207680
|
0.742
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr6_-_46293628
|
0.740
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr14_-_64006402
|
0.734
|
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform
|
|
chr4_-_186577858
|
0.732
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_131132690
|
0.730
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr5_-_56778553
|
0.728
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chrX_+_102862833
|
0.724
|
NM_001006933
NM_032926
|
TCEAL3
|
transcription elongation factor A (SII)-like 3
|
|
chr16_+_6069094
|
0.723
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr18_+_32073343
|
0.719
|
|
DTNA
|
dystrobrevin, alpha
|
|
chr3_-_195310985
|
0.717
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr20_+_55904814
|
0.698
|
NM_012444
NM_198265
|
SPO11
|
SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae)
|
|
chr19_-_7698569
|
0.697
|
NM_174895
|
PCP2
|
Purkinje cell protein 2
|
|
chr4_-_76957139
|
0.692
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr19_+_52848681
|
0.688
|
NM_173530
|
ZNF610
|
zinc finger protein 610
|
|
chr12_-_8815479
|
0.686
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr10_-_131762017
|
0.680
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr12_+_27849427
|
0.675
|
NM_001029874
|
REP15
|
RAB15 effector protein
|
|
chr4_-_186578122
|
0.669
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_55559749
|
0.663
|
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr10_+_50507186
|
0.663
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr3_+_158991090
|
0.662
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr20_+_2517252
|
0.660
|
NM_080751
|
TMC2
|
transmembrane channel-like 2
|
|
chr1_+_145472454
|
0.660
|
|
ANKRD34A
|
ankyrin repeat domain 34A
|
|
chr2_+_27498288
|
0.659
|
NM_173650
|
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma
|
|
chr17_-_9929567
|
0.656
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr7_-_28220342
|
0.648
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr2_-_100722328
|
0.647
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr11_-_84028379
|
0.647
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_+_150245182
|
0.646
|
NM_024579
|
C1orf54
|
chromosome 1 open reading frame 54
|
|
chr3_+_107096187
|
0.642
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr3_+_50654868
|
0.636
|
|
MAPKAPK3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr4_+_75174189
|
0.630
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr8_+_21900389
|
0.621
|
NM_003867
|
FGF17
|
fibroblast growth factor 17
|
|
chr12_+_21525801
|
0.620
|
NM_000415
|
IAPP
|
islet amyloid polypeptide
|
|
chr5_-_148442606
|
0.618
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr8_-_107782417
|
0.616
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr15_+_75105152
|
0.614
|
NM_021819
|
LMAN1L
|
lectin, mannose-binding, 1 like
|
|
chr2_-_16081703
|
0.611
|
|
MYCNOS
|
MYCN opposite strand/antisense RNA (non-protein coding)
|
|
chr18_-_3219846
|
0.610
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr7_+_136553398
|
0.609
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr5_-_131132709
|
0.605
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr3_-_108672547
|
0.603
|
NM_005459
|
GUCA1C
|
guanylate cyclase activator 1C
|
|
chr14_+_21467413
|
0.599
|
NM_014579
|
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2
|
|
chr12_-_8815266
|
0.598
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr18_+_32073245
|
0.595
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr7_+_129847699
|
0.592
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr2_+_233404436
|
0.581
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chr17_+_32612686
|
0.581
|
NM_002986
|
CCL11
|
chemokine (C-C motif) ligand 11
|
|
chr1_-_241799101
|
0.574
|
NM_001821
|
CHML
|
choroideremia-like (Rab escort protein 2)
|
|
chr5_-_131132642
|
0.572
|
|
FNIP1
|
folliculin interacting protein 1
|
|
chr7_-_99766191
|
0.571
|
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr3_-_71542628
|
0.567
|
|
FOXP1
|
forkhead box P1
|
|
chr20_+_138121
|
0.565
|
NM_139074
|
DEFB127
|
defensin, beta 127
|
|
chr17_+_25799024
|
0.565
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr5_-_132272803
|
0.562
|
|
|
|
|
chr2_-_40657419
|
0.559
|
NM_001112800
NM_001112801
NM_001252624
NM_021097
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr5_+_140593508
|
0.553
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr8_+_24771276
|
0.552
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr5_+_61714710
|
0.550
|
NM_001134779
|
IPO11
|
importin 11
|
|
chr1_-_35370983
|
0.546
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr1_+_160051343
|
0.544
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr16_+_30386097
|
0.544
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr8_-_22089850
|
0.543
|
NM_001099335
NM_014759
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr8_+_24771268
|
0.542
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_+_48516428
|
0.542
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chr19_-_11565612
|
0.540
|
|
|
|
|
chr12_+_26348268
|
0.539
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr7_-_80548322
|
0.539
|
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr3_+_158991035
|
0.538
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr3_-_139396773
|
0.538
|
NM_001200047
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr7_+_143013218
|
0.535
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr12_-_26277807
|
0.531
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr22_-_38379790
|
0.527
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chrX_+_122318095
|
0.525
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr2_-_217724781
|
0.525
|
NM_003284
|
TNP1
|
transition protein 1 (during histone to protamine replacement)
|
|
chr15_+_78556485
|
0.524
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr11_+_6280903
|
0.521
|
NM_176875
|
CCKBR
|
cholecystokinin B receptor
|
|
chr7_+_107220622
|
0.518
|
|
BCAP29
|
B-cell receptor-associated protein 29
|
|
chr17_-_39041436
|
0.517
|
NM_019010
|
KRT20
|
keratin 20
|
|
chr9_-_104500217
|
0.515
|
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chr11_-_64703334
|
0.514
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr9_+_109686436
|
0.511
|
|
ZNF462
|
zinc finger protein 462
|
|
chrX_-_152938742
|
0.509
|
NM_001135740
|
PNCK
|
pregnancy up-regulated non-ubiquitously expressed CaM kinase
|
|
chr12_-_111180499
|
0.508
|
|
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme
|