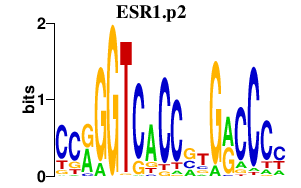
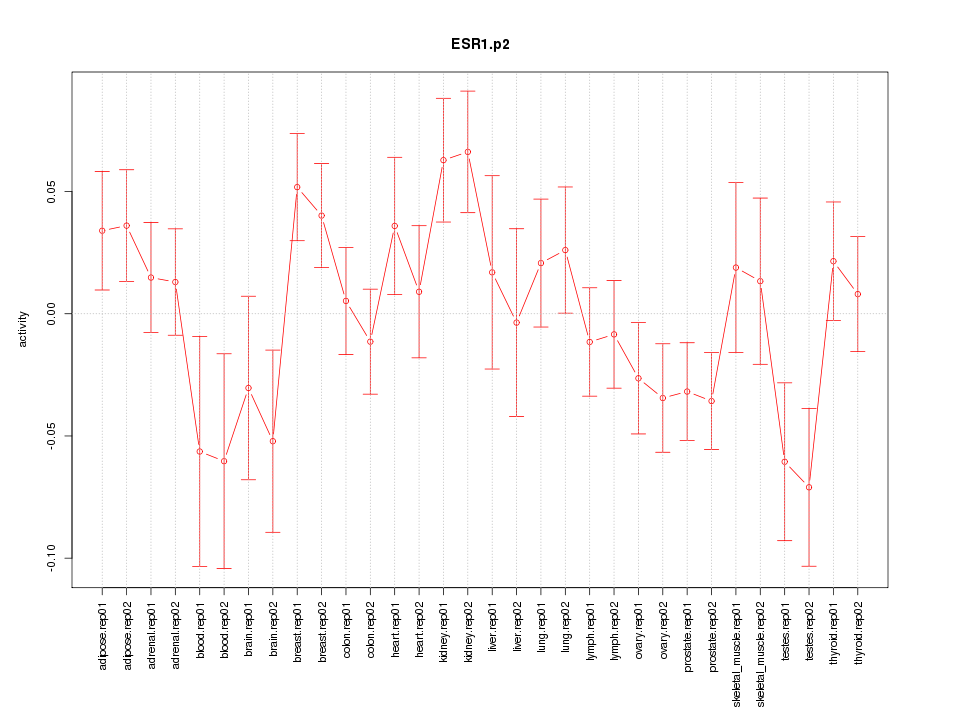
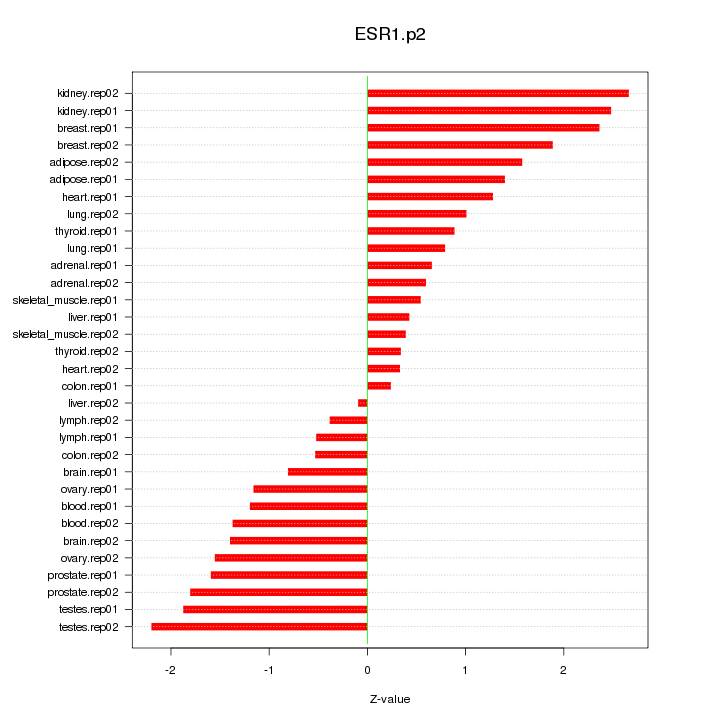
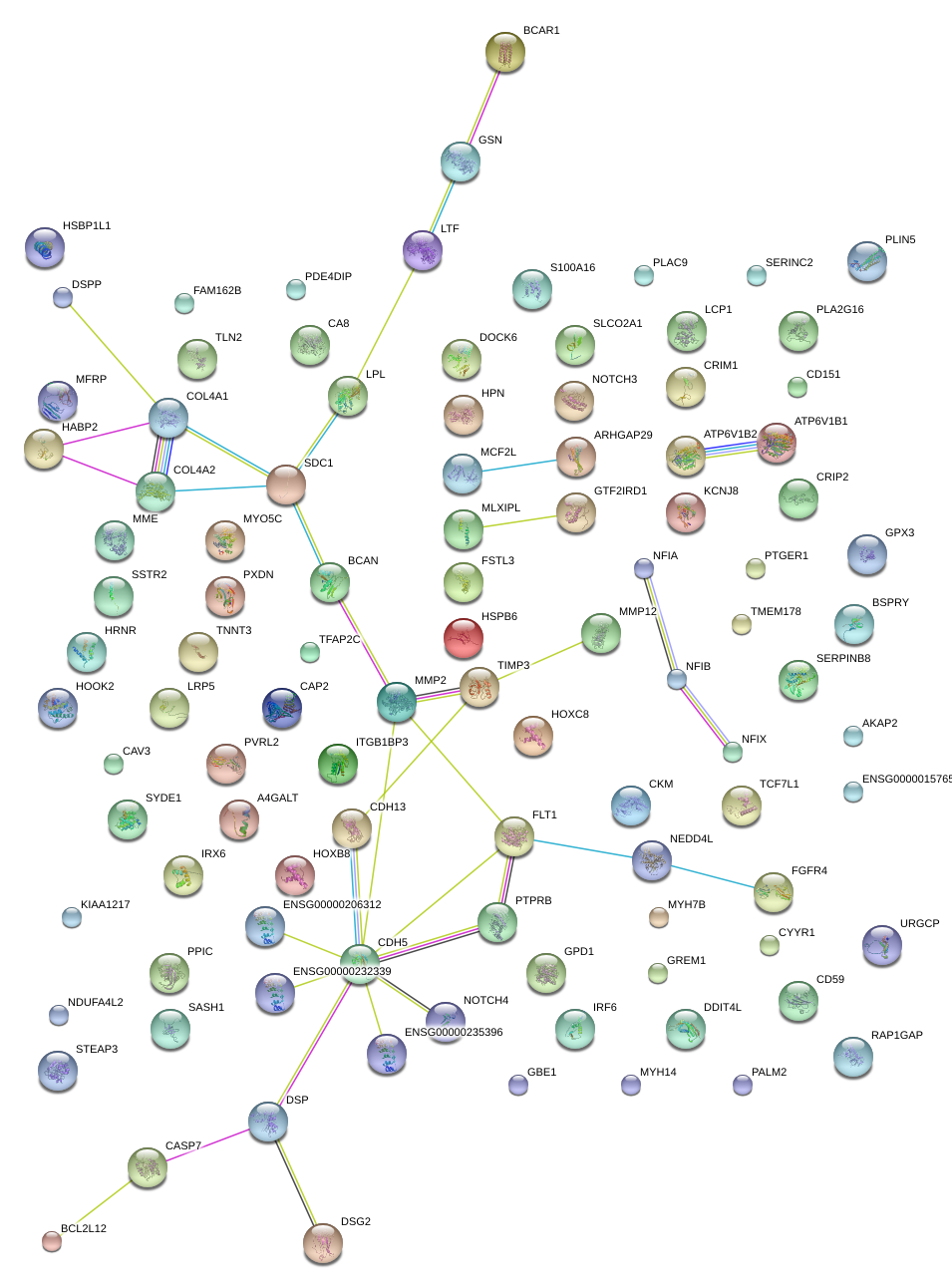
|
chr5_+_150400132
|
2.207
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr22_-_43116796
|
2.074
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr3_-_81810802
|
1.924
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr13_+_110959592
|
1.899
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr3_-_81810816
|
1.847
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr22_-_43117270
|
1.842
|
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr3_-_133748727
|
1.836
|
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr13_-_110959441
|
1.828
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_+_36582812
|
1.826
|
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr3_-_133748828
|
1.805
|
NM_005630
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1
|
|
chr3_+_154797435
|
1.788
|
NM_000902
|
MME
|
membrane metallo-endopeptidase
|
|
chr3_-_81810796
|
1.757
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr3_-_81810618
|
1.672
|
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr5_+_150399980
|
1.647
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr19_+_15218141
|
1.616
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr20_+_55204307
|
1.609
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr6_+_148663728
|
1.540
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr6_+_148663953
|
1.528
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr14_-_106714594
|
1.480
|
|
|
|
|
chr16_-_75285383
|
1.477
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr22_+_33197678
|
1.441
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr22_+_33197744
|
1.410
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr19_+_45349560
|
1.343
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr18_+_77724575
|
1.327
|
NM_001136180
|
HSBP1L1
|
heat shock factor binding protein 1-like 1
|
|
chr11_+_68079999
|
1.318
|
NM_002335
|
LRP5
|
low density lipoprotein receptor-related protein 5
|
|
chr5_-_122372413
|
1.317
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr16_+_55358470
|
1.313
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr10_+_23983674
|
1.311
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr1_+_31886659
|
1.280
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr7_-_73038814
|
1.280
|
NM_032951
NM_032952
NM_032953
NM_032954
|
MLXIPL
|
MLX interacting protein-like
|
|
chr1_+_31885962
|
1.256
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr10_+_115310589
|
1.255
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr14_+_105941061
|
1.247
|
NM_001312
|
CRIP2
|
cysteine-rich protein 2
|
|
chr10_+_115438934
|
1.246
|
NM_033340
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr19_-_36247917
|
1.204
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr12_+_54402795
|
1.189
|
NM_022658
|
HOXC8
|
homeobox C8
|
|
chr19_+_676346
|
1.174
|
NM_005860
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein)
|
|
chr15_+_62682682
|
1.170
|
|
TLN2
|
talin 2
|
|
chr12_+_50497790
|
1.150
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr11_+_832965
|
1.148
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr2_+_71162995
|
1.119
|
NM_001692
|
ATP6V1B1
|
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1
|
|
chr18_+_29078026
|
1.103
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr3_-_46505077
|
1.099
|
NM_001199149
|
LTF
|
lactotransferrin
|
|
chr16_+_82660573
|
1.092
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr12_-_71031174
|
1.076
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr2_+_119981389
|
1.072
|
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr10_+_81892257
|
1.072
|
NM_001012973
|
PLAC9
|
placenta-specific 9
|
|
chr5_+_176513819
|
1.065
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr18_+_55711388
|
1.063
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_+_66400524
|
1.055
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr19_+_3933100
|
1.054
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr16_+_55515468
|
1.052
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_-_153585504
|
1.042
|
NM_080388
|
S100A16
|
S100 calcium binding protein A16
|
|
chr10_+_81892429
|
1.033
|
|
PLAC9
|
placenta-specific 9
|
|
chr13_+_113741668
|
1.030
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr8_+_19796840
|
1.028
|
|
LPL
|
lipoprotein lipase
|
|
chr19_-_45826232
|
1.024
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr18_+_55711890
|
1.021
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr19_+_35550794
|
1.021
|
|
HPN
|
hepsin
|
|
chr11_-_119211034
|
1.017
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr19_-_11373117
|
1.017
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr19_-_15311773
|
1.016
|
NM_000435
|
NOTCH3
|
notch 3
|
|
chr19_-_12886263
|
1.012
|
|
HOOK2
|
hook homolog 2 (Drosophila)
|
|
chr2_+_39893021
|
1.008
|
NM_152390
|
TMEM178
|
transmembrane protein 178
|
|
chr6_+_7541863
|
1.000
|
NM_001008844
NM_004415
|
DSP
|
desmoplakin
|
|
chr19_+_35532617
|
0.999
|
|
HPN
|
hepsin
|
|
chr8_+_19796929
|
0.999
|
|
LPL
|
lipoprotein lipase
|
|
chr11_+_1940798
|
0.997
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr6_+_148664124
|
0.996
|
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr6_+_7541837
|
0.988
|
|
DSP
|
desmoplakin
|
|
chr10_+_81892497
|
0.986
|
|
PLAC9
|
placenta-specific 9
|
|
chr12_-_57634474
|
0.980
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr19_-_4535202
|
0.980
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr3_+_8775485
|
0.954
|
NM_001234
NM_033337
|
CAV3
|
caveolin 3
|
|
chr11_-_33757872
|
0.951
|
NM_000611
NM_203329
NM_203330
NM_203331
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr21_-_27945267
|
0.950
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr6_+_7541892
|
0.941
|
|
DSP
|
desmoplakin
|
|
chr1_-_145039631
|
0.931
|
NM_001198832
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr19_+_35532640
|
0.923
|
|
HPN
|
hepsin
|
|
chr2_-_20425166
|
0.922
|
NM_001006946
|
SDC1
|
syndecan 1
|
|
chr13_-_29069215
|
0.919
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr2_+_85360373
|
0.917
|
NM_031283
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box)
|
|
chr19_+_50706868
|
0.912
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr9_-_14313509
|
0.910
|
|
NFIB
|
nuclear factor I/B
|
|
chr13_+_113622758
|
0.901
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr2_-_1748240
|
0.894
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr12_-_21927614
|
0.887
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr19_-_14586122
|
0.886
|
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr9_+_112810877
|
0.871
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr19_-_45826131
|
0.866
|
|
CKM
|
creatine kinase, muscle
|
|
chr1_-_94702916
|
0.865
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr17_-_46692300
|
0.861
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr2_-_1748317
|
0.859
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr7_+_73868111
|
0.851
|
NM_001199207
NM_005685
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr1_-_21978287
|
0.849
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr16_+_66400559
|
0.846
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr2_+_119981383
|
0.835
|
NM_001008410
NM_018234
NM_182915
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr1_-_209979388
|
0.833
|
NM_001206696
NM_006147
|
IRF6
|
interferon regulatory factor 6
|
|
chr15_+_33010174
|
0.830
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr17_+_71161110
|
0.817
|
NM_001050
|
SSTR2
|
somatostatin receptor 2
|
|
chr6_-_117086812
|
0.813
|
NM_001085480
|
FAM162B
|
family with sequence similarity 162, member B
|
|
chr19_+_45349584
|
0.811
|
|
PVRL2
|
poliovirus receptor-related 2 (herpesvirus entry mediator B)
|
|
chr9_+_123963728
|
0.808
|
|
GSN
|
gelsolin
|
|
chr15_-_52587851
|
0.803
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr8_-_61193882
|
0.802
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr9_+_116111811
|
0.799
|
NM_017688
|
BSPRY
|
B-box and SPRY domain containing
|
|
chr11_-_63381840
|
0.790
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr4_-_101111606
|
0.783
|
NM_145244
|
DDIT4L
|
DNA-damage-inducible transcript 4-like
|
|
chr6_+_17393610
|
0.781
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr11_-_72353435
|
0.778
|
NM_001143839
NM_001243784
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr4_+_78978723
|
0.778
|
NM_001166133
NM_025074
|
FRAS1
|
Fraser syndrome 1
|
|
chr14_+_100203998
|
0.777
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr9_+_112810949
|
0.770
|
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr6_-_154831704
|
0.768
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr11_-_118023534
|
0.763
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr22_-_38794853
|
0.761
|
|
LOC400927
|
TPTE and PTEN homologous inositol lipid phosphatase pseudogene
|
|
chr9_+_4490193
|
0.758
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr9_+_87283453
|
0.757
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr7_-_134855237
|
0.755
|
NM_001243752
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr11_-_63381907
|
0.754
|
NM_007069
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr11_-_1785190
|
0.751
|
NM_001909
|
CTSD
|
cathepsin D
|
|
chr19_-_14586163
|
0.747
|
NM_000955
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa
|
|
chr9_+_129376721
|
0.747
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr17_-_18266691
|
0.744
|
NM_004169
NM_148918
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble)
|
|
chr12_+_6419601
|
0.741
|
NM_018173
|
PLEKHG6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6
|
|
chr9_-_86153048
|
0.739
|
NM_001244959
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr3_+_154797644
|
0.737
|
NM_007287
|
MME
|
membrane metallo-endopeptidase
|
|
chr4_-_8073711
|
0.736
|
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr13_+_51796363
|
0.730
|
NM_001242312
NM_145019
|
FAM124A
|
family with sequence similarity 124A
|
|
chr19_-_49576120
|
0.727
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr4_-_41216455
|
0.712
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr13_+_76210449
|
0.711
|
|
LMO7
|
LIM domain 7
|
|
chr9_-_136150629
|
0.710
|
NM_020469
|
ABO
|
ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase)
|
|
chr1_-_94703252
|
0.706
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr9_+_137533643
|
0.706
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr7_+_73868236
|
0.704
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr8_-_17658385
|
0.696
|
NM_001001924
NM_001001925
|
MTUS1
|
microtubule associated tumor suppressor 1
|
|
chr1_+_55446464
|
0.696
|
NM_182532
|
TMEM61
|
transmembrane protein 61
|
|
chr19_+_840970
|
0.694
|
NM_002777
|
PRTN3
|
proteinase 3
|
|
chr3_-_81810914
|
0.693
|
NM_000158
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1
|
|
chr3_+_69788562
|
0.693
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr4_-_57976550
|
0.690
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr5_-_524444
|
0.690
|
NM_004174
|
SLC9A3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3
|
|
chr16_+_23313590
|
0.689
|
NM_000336
|
SCNN1B
|
sodium channel, nonvoltage-gated 1, beta
|
|
chr19_-_19384022
|
0.688
|
NM_001001524
|
TM6SF2
|
transmembrane 6 superfamily member 2
|
|
chr13_+_103451398
|
0.686
|
NM_001159596
NM_017693
|
BIVM
|
basic, immunoglobulin-like variable motif containing
|
|
chr17_-_48264039
|
0.683
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr17_-_48266572
|
0.678
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr6_+_147829784
|
0.673
|
NM_001030060
|
SAMD5
|
sterile alpha motif domain containing 5
|
|
chr11_-_63381485
|
0.670
|
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr19_+_3585541
|
0.669
|
NM_133261
|
GIPC3
|
GIPC PDZ domain containing family, member 3
|
|
chr1_-_19615203
|
0.667
|
NM_012067
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase)
|
|
chr11_-_2019007
|
0.658
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr13_+_25254548
|
0.658
|
NM_001185085
NM_001676
|
ATP12A
|
ATPase, H+/K+ transporting, nongastric, alpha polypeptide
|
|
chr9_-_33402505
|
0.654
|
NM_001170
|
AQP7
|
aquaporin 7
|
|
chr17_+_34058678
|
0.651
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chrX_+_135229536
|
0.650
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr22_+_50919970
|
0.647
|
NM_024866
NM_001253845
|
ADM2
|
adrenomedullin 2
|
|
chr3_+_171757413
|
0.645
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr1_-_68299047
|
0.643
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr20_-_45280059
|
0.643
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr2_-_1748285
|
0.642
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr20_+_62372436
|
0.642
|
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr19_-_46272102
|
0.640
|
|
|
|
|
chr16_-_57836380
|
0.640
|
NM_001130100
NM_005550
|
KIFC3
|
kinesin family member C3
|
|
chr1_-_27240430
|
0.639
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr11_-_1785104
|
0.637
|
|
CTSD
|
cathepsin D
|
|
chr1_+_43148048
|
0.635
|
NM_004559
|
YBX1
|
Y box binding protein 1
|
|
chr8_-_25315791
|
0.632
|
|
KCTD9
|
potassium channel tetramerisation domain containing 9
|
|
chr8_+_21924369
|
0.624
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr18_+_55711746
|
0.624
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr3_-_46735133
|
0.624
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr22_+_44319685
|
0.622
|
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr6_-_154831652
|
0.620
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr2_-_216946463
|
0.620
|
NM_018441
|
PECR
|
peroxisomal trans-2-enoyl-CoA reductase
|
|
chr21_+_42540092
|
0.614
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr22_+_19159198
|
0.613
|
|
LOC100652736
|
uncharacterized LOC100652736
|
|
chr22_-_51016343
|
0.612
|
NM_001145137
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr6_+_46620631
|
0.611
|
NM_001204051
NM_001204052
NM_004277
|
SLC25A27
|
solute carrier family 25, member 27
|
|
chr8_-_10587942
|
0.609
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr5_-_180076544
|
0.608
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr16_-_57836370
|
0.604
|
|
KIFC3
|
kinesin family member C3
|
|
chr22_-_19929322
|
0.601
|
|
TXNRD2
|
thioredoxin reductase 2
|
|
chr6_-_154831788
|
0.600
|
|
CNKSR3
|
CNKSR family member 3
|
|
chr11_+_833007
|
0.596
|
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr22_+_44319618
|
0.595
|
NM_025225
|
PNPLA3
|
patatin-like phospholipase domain containing 3
|
|
chr12_-_56122124
|
0.594
|
|
CD63
|
CD63 molecule
|
|
chr11_+_832909
|
0.592
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr1_+_43148098
|
0.592
|
|
YBX1
|
Y box binding protein 1
|
|
chr19_+_49660997
|
0.591
|
NM_001195227
NM_017636
|
TRPM4
|
transient receptor potential cation channel, subfamily M, member 4
|
|
chr8_+_38854431
|
0.590
|
NM_003816
|
ADAM9
|
ADAM metallopeptidase domain 9
|
|
chr11_-_63381940
|
0.589
|
NM_001128203
|
PLA2G16
|
phospholipase A2, group XVI
|
|
chr16_+_23194037
|
0.589
|
NM_001039
|
SCNN1G
|
sodium channel, nonvoltage-gated 1, gamma
|
|
chr7_-_134855383
|
0.589
|
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr12_+_52306112
|
0.588
|
NM_001077401
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr1_+_159750756
|
0.587
|
NM_017823
|
DUSP23
|
dual specificity phosphatase 23
|
|
chr1_-_59042827
|
0.587
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr21_+_42540054
|
0.587
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr16_-_54320328
|
0.585
|
|
IRX3
|
iroquois homeobox 3
|
|
chr9_-_112260529
|
0.582
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|