|
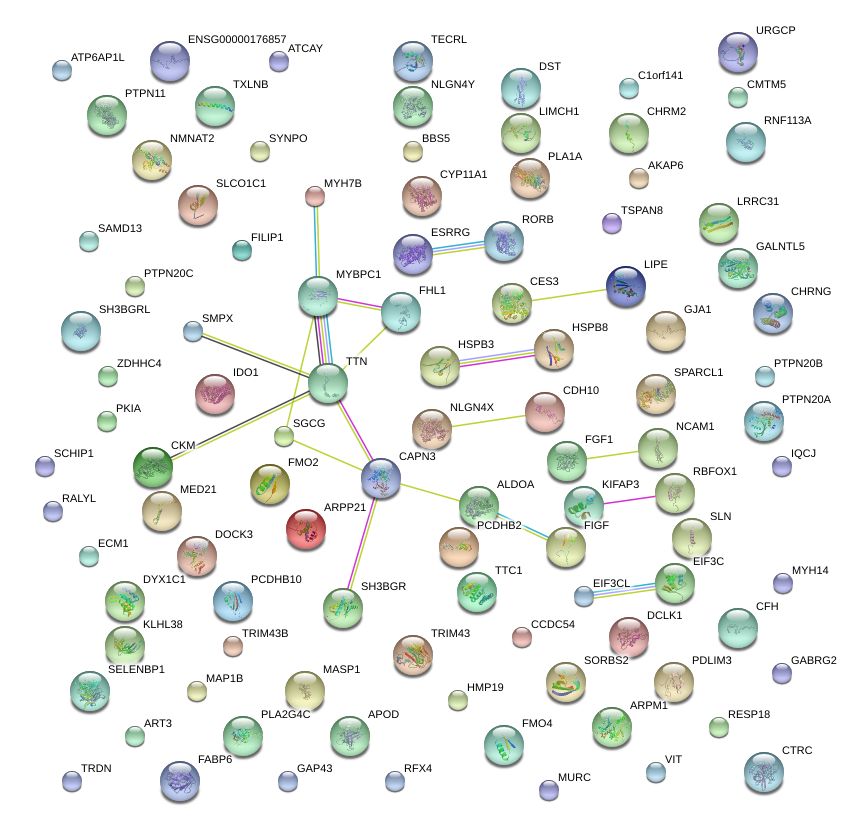
chr13_+_23755059
|
15.475
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr7_+_151653463
|
11.754
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr12_+_101988746
|
8.562
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr5_+_53751430
|
8.291
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr16_+_6823809
|
5.915
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr16_+_30064470
|
5.687
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr12_+_112890999
|
5.251
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr16_+_30064410
|
4.924
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr14_+_32798478
|
4.601
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr19_-_48614013
|
4.530
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr4_-_186456580
|
4.513
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr1_+_150480486
|
4.316
|
NM_001202858
NM_004425
NM_022664
|
ECM1
|
extracellular matrix protein 1
|
|
chr5_+_173472692
|
4.277
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr3_+_158787040
|
4.231
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr19_-_45822841
|
4.088
|
|
CKM
|
creatine kinase, muscle
|
|
chr14_+_23845932
|
4.020
|
NM_001037288
NM_138460
|
CMTM5
|
CKLF-like MARVEL transmembrane domain containing 5
|
|
chr5_+_173472737
|
3.875
|
|
HMP19
|
HMP19 protein
|
|
chr4_-_65275000
|
3.756
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr6_-_123957941
|
3.720
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr13_-_36429798
|
3.674
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr2_-_179672056
|
3.650
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chrX_-_21776158
|
3.632
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr1_+_84764048
|
3.622
|
NM_001010971
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr5_+_161494647
|
3.609
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr3_-_195310770
|
3.568
|
|
APOD
|
apolipoprotein D
|
|
chr15_+_42696957
|
3.454
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr5_+_159614373
|
3.369
|
NM_001130958
|
FABP6
|
fatty acid binding protein 6, ileal
|
|
chr3_-_195310985
|
3.369
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr11_+_112832089
|
3.286
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr9_+_103340335
|
3.214
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr7_+_136553398
|
3.109
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr3_+_119316694
|
3.108
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr10_-_48806862
|
3.085
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr8_+_85095501
|
3.058
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr3_-_169487602
|
3.040
|
NM_032487
|
ARPM1
|
actin related protein M1
|
|
chr3_-_12586962
|
2.997
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr15_-_74658498
|
2.967
|
NM_001099773
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr12_+_20848288
|
2.936
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr1_-_67594219
|
2.904
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr5_+_149997205
|
2.819
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr10_-_46619935
|
2.811
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr1_+_84768333
|
2.806
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr4_+_76995862
|
2.801
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr3_+_35721115
|
2.782
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_+_6617035
|
2.770
|
NM_001134387
NM_001134388
NM_001134389
NM_018106
|
ZDHHC4
|
zinc finger, DHHC-type containing 4
|
|
chr16_+_28699878
|
2.759
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr3_+_107096187
|
2.733
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr1_+_171154387
|
2.725
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr6_-_139613114
|
2.710
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr2_+_233404436
|
2.707
|
NM_005199
|
CHRNG
|
cholinergic receptor, nicotinic, gamma
|
|
chr19_-_42931500
|
2.651
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr5_+_81601165
|
2.639
|
NM_001017971
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like
|
|
chr4_+_41614725
|
2.628
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_186577858
|
2.618
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_24645084
|
2.606
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr1_-_216978708
|
2.583
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr12_-_71551564
|
2.547
|
|
TSPAN8
|
tetraspanin 8
|
|
chr9_+_77112251
|
2.505
|
NM_006914
|
RORB
|
RAR-related orphan receptor B
|
|
chr2_+_170335948
|
2.496
|
NM_152384
|
BBS5
|
Bardet-Biedl syndrome 5
|
|
chrX_-_15402469
|
2.493
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr8_-_124665165
|
2.482
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chrX_+_135278912
|
2.470
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr1_-_170043608
|
2.459
|
NM_001204514
NM_001204516
NM_014970
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr1_+_15764934
|
2.446
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr12_+_27175501
|
2.445
|
|
MED21
|
mediator complex subunit 21
|
|
chr12_+_27175482
|
2.436
|
NM_004264
|
MED21
|
mediator complex subunit 21
|
|
chr21_+_40823779
|
2.413
|
NM_007341
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chr1_-_183274008
|
2.393
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr12_+_107078476
|
2.361
|
NM_032491
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr6_+_121756744
|
2.355
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr4_-_186578122
|
2.339
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_115342150
|
2.337
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr12_-_71551778
|
2.336
|
NM_004616
|
TSPAN8
|
tetraspanin 8
|
|
chr6_-_56507585
|
2.324
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr15_-_55800431
|
2.312
|
NM_001033559
NM_001033560
NM_130810
|
DYX1C1
|
dyslexia susceptibility 1 candidate 1
|
|
chr5_+_140566655
|
2.293
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr4_-_88450601
|
2.292
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr5_+_159436143
|
2.254
|
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr19_+_50796820
|
2.245
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr5_+_140474198
|
2.229
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr3_-_169587598
|
2.217
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr1_-_151345156
|
2.213
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr8_+_79428335
|
2.208
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr5_+_71502700
|
2.178
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr19_+_3880675
|
2.152
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr8_+_85095452
|
2.143
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr12_+_119616594
|
2.124
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr16_+_67001377
|
2.110
|
NM_001185176
|
CES3
|
carboxylesterase 3
|
|
chr2_+_36923902
|
2.072
|
|
VIT
|
vitrin
|
|
chr16_-_28437663
|
2.058
|
NM_001199142
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C
|
|
chr6_-_76203256
|
2.038
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr21_+_40817793
|
2.020
|
NM_001001713
|
SH3BGR
|
SH3 domain binding glutamic acid-rich protein
|
|
chrX_-_6146655
|
1.999
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr2_-_220197898
|
1.995
|
NM_001007089
|
RESP18
|
regulated endocrine-specific protein 18 homolog (rat)
|
|
chr3_-_187009469
|
1.933
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr11_-_107582733
|
1.932
|
NM_003063
|
SLN
|
sarcolipin
|
|
chr5_+_159436155
|
1.929
|
NM_003314
|
TTC1
|
tetratricopeptide repeat domain 1
|
|
chr8_+_39771327
|
1.920
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr5_-_142077508
|
1.913
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_50712671
|
1.913
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr2_-_96150478
|
1.908
|
NM_001164464
|
TRIM43B
|
tripartite motif containing 43B
|
|
chr8_-_52721903
|
1.896
|
NM_144651
|
PXDNL
|
peroxidasin homolog (Drosophila)-like
|
|
chr1_-_163172863
|
1.860
|
NM_003617
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr1_-_170038055
|
1.855
|
NM_001204517
|
KIFAP3
|
kinesin-associated protein 3
|
|
chr11_+_113931228
|
1.849
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr19_+_3880660
|
1.829
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr19_+_35629727
|
1.818
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr1_+_172389827
|
1.815
|
NM_139240
|
C1orf105
|
chromosome 1 open reading frame 105
|
|
chr9_-_71590754
|
1.813
|
|
|
|
|
chr12_-_15038724
|
1.807
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr5_+_140571903
|
1.801
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr10_-_45474210
|
1.796
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr18_+_32073245
|
1.778
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_-_72374919
|
1.768
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr12_-_71148383
|
1.762
|
NM_130846
NM_001207016
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_-_176057324
|
1.762
|
NM_001001502
NM_003085
|
SNCB
|
synuclein, beta
|
|
chr1_-_153599523
|
1.759
|
NM_001024213
|
S100A13
|
S100 calcium binding protein A13
|
|
chr11_+_114549199
|
1.757
|
NM_182495
|
FAM55B
|
family with sequence similarity 55, member B
|
|
chr10_-_97200895
|
1.734
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr7_+_86273219
|
1.720
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr16_-_23724820
|
1.711
|
NM_033266
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2
|
|
chr11_-_790103
|
1.708
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr17_+_44101549
|
1.700
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr4_+_154178525
|
1.697
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr3_+_158991090
|
1.682
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_-_8424217
|
1.662
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr19_+_52873169
|
1.661
|
NM_001145434
|
ZNF880
|
zinc finger protein 880
|
|
chr12_+_124773709
|
1.654
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr6_+_101846668
|
1.654
|
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr4_+_166794409
|
1.654
|
NM_001204760
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr6_-_105584982
|
1.641
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr10_-_115423640
|
1.636
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr6_+_150263120
|
1.636
|
NM_025217
|
ULBP2
|
UL16 binding protein 2
|
|
chr12_-_111358353
|
1.635
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr4_+_119809978
|
1.630
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr3_+_135741576
|
1.630
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr9_-_19786886
|
1.626
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr1_-_168698412
|
1.614
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr9_+_113431050
|
1.613
|
NM_001166280
NM_001166281
NM_005592
|
MUSK
|
muscle, skeletal, receptor tyrosine kinase
|
|
chr3_+_11267666
|
1.612
|
NM_001098211
|
HRH1
|
histamine receptor H1
|
|
chr4_+_166299942
|
1.609
|
NM_001873
|
CPE
|
carboxypeptidase E
|
|
chr3_-_58563070
|
1.603
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr5_+_78985645
|
1.602
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chr14_+_51026742
|
1.601
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr6_-_112575708
|
1.600
|
|
LAMA4
|
laminin, alpha 4
|
|
chr19_+_9296269
|
1.591
|
NM_175883
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2
|
|
chr1_-_151777741
|
1.590
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr4_-_70826712
|
1.581
|
NM_001891
|
CSN2
|
casein beta
|
|
chr19_+_30863318
|
1.563
|
NM_014717
|
ZNF536
|
zinc finger protein 536
|
|
chr10_-_7659186
|
1.558
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr11_+_120195837
|
1.554
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr6_-_112575766
|
1.554
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr19_-_5846465
|
1.546
|
|
FUT3
|
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
|
|
chr20_+_10199455
|
1.546
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr6_+_44244465
|
1.537
|
|
TMEM151B
|
transmembrane protein 151B
|
|
chr8_-_82359620
|
1.525
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr5_+_140220811
|
1.521
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr3_+_63428752
|
1.519
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr20_-_44538654
|
1.512
|
|
PLTP
|
phospholipid transfer protein
|
|
chr6_-_53530505
|
1.505
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr2_+_207406815
|
1.503
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chrX_+_8432870
|
1.502
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr5_+_140529619
|
1.491
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr19_+_3880617
|
1.461
|
NM_033064
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr14_+_79745653
|
1.453
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr16_+_8715473
|
1.453
|
NM_024109
|
METTL22
|
methyltransferase like 22
|
|
chr6_+_101846860
|
1.449
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr6_+_69344939
|
1.446
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr4_+_113739203
|
1.444
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr15_+_78558522
|
1.434
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_-_41166336
|
1.433
|
NM_133639
|
RHOV
|
ras homolog gene family, member V
|
|
chr11_+_112831968
|
1.425
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr10_-_69597768
|
1.424
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr16_+_30034654
|
1.421
|
NM_001109659
NM_001109660
|
C16orf92
|
chromosome 16 open reading frame 92
|
|
chr2_+_36923832
|
1.405
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr7_-_87849341
|
1.403
|
|
SRI
|
sorcin
|
|
chr11_+_57310113
|
1.390
|
NM_001105565
|
SMTNL1
|
smoothelin-like 1
|
|
chr18_+_28898051
|
1.383
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr1_+_77333158
|
1.383
|
NM_030965
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5
|
|
chr11_+_60260250
|
1.373
|
NM_001164470
NM_017716
|
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12
|
|
chr1_+_74701070
|
1.371
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chrX_-_6145887
|
1.369
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr9_-_95298251
|
1.363
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chrX_-_44202834
|
1.363
|
NM_025184
|
EFHC2
|
EF-hand domain (C-terminal) containing 2
|
|
chr22_-_43036606
|
1.363
|
NM_001165877
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2
|
|
chr7_+_121513158
|
1.350
|
NM_001206838
NM_001206839
NM_002851
|
PTPRZ1
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1
|
|
chr5_+_175308843
|
1.339
|
|
CPLX2
|
complexin 2
|
|
chr12_-_22089627
|
1.337
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr9_+_102677457
|
1.336
|
|
STX17
|
syntaxin 17
|
|
chr22_-_38840032
|
1.332
|
NM_004981
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr19_+_50691437
|
1.324
|
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr5_-_10308167
|
1.324
|
NM_138809
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas)
|
|
chr17_+_3377346
|
1.323
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr13_-_61989209
|
1.322
|
|
PCDH20
|
protocadherin 20
|
|
chr9_+_71944240
|
1.321
|
NM_004816
|
FAM189A2
|
family with sequence similarity 189, member A2
|
|
chr2_-_211168315
|
1.319
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr22_-_46659218
|
1.313
|
NM_006071
|
PKDREJ
|
polycystic kidney disease (polycystin) and REJ homolog (sperm receptor for egg jelly homolog, sea urchin)
|
|
chr16_-_31439650
|
1.311
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr21_-_43346780
|
1.310
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr7_+_80267782
|
1.310
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|