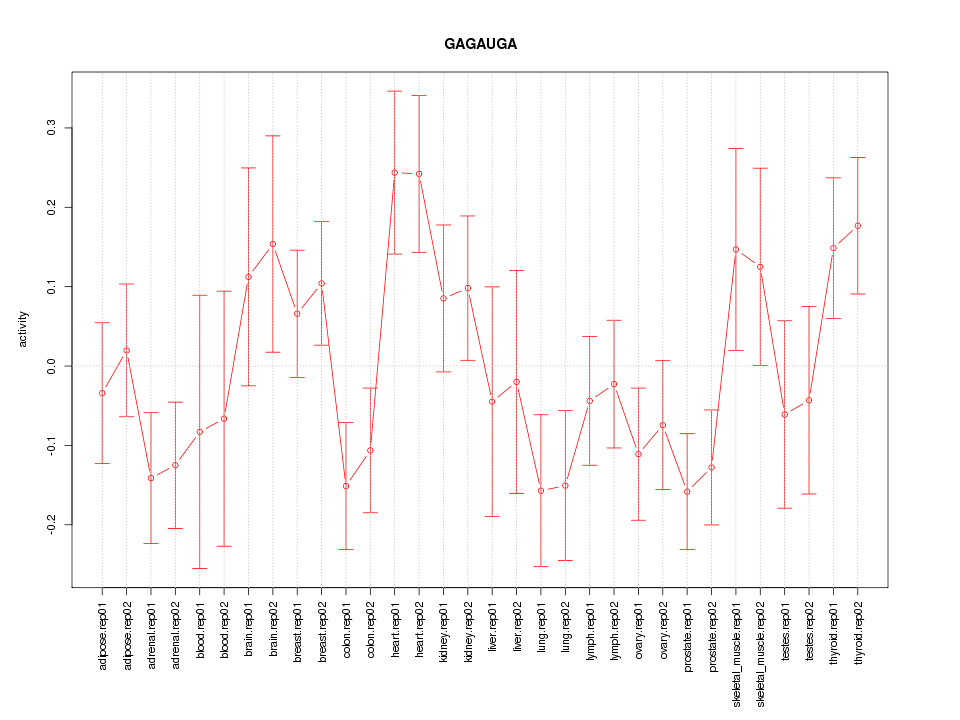
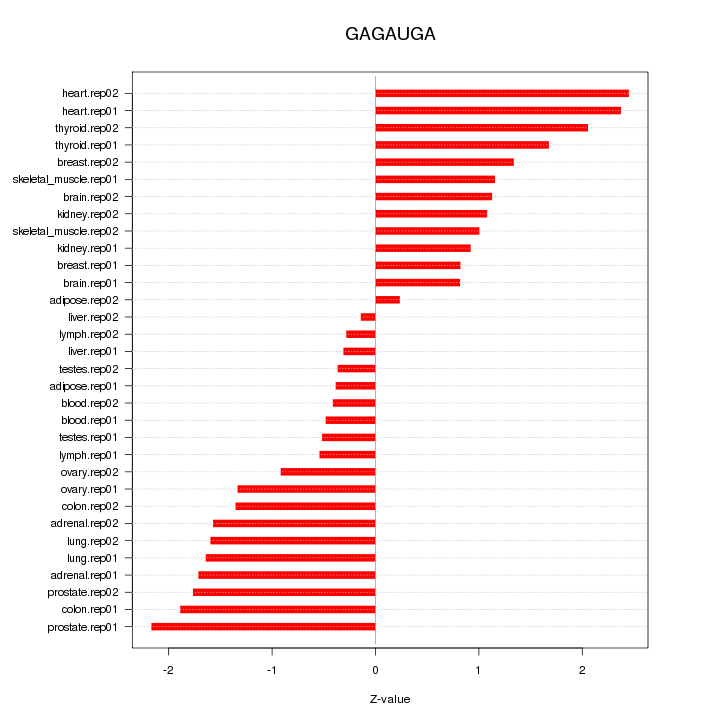
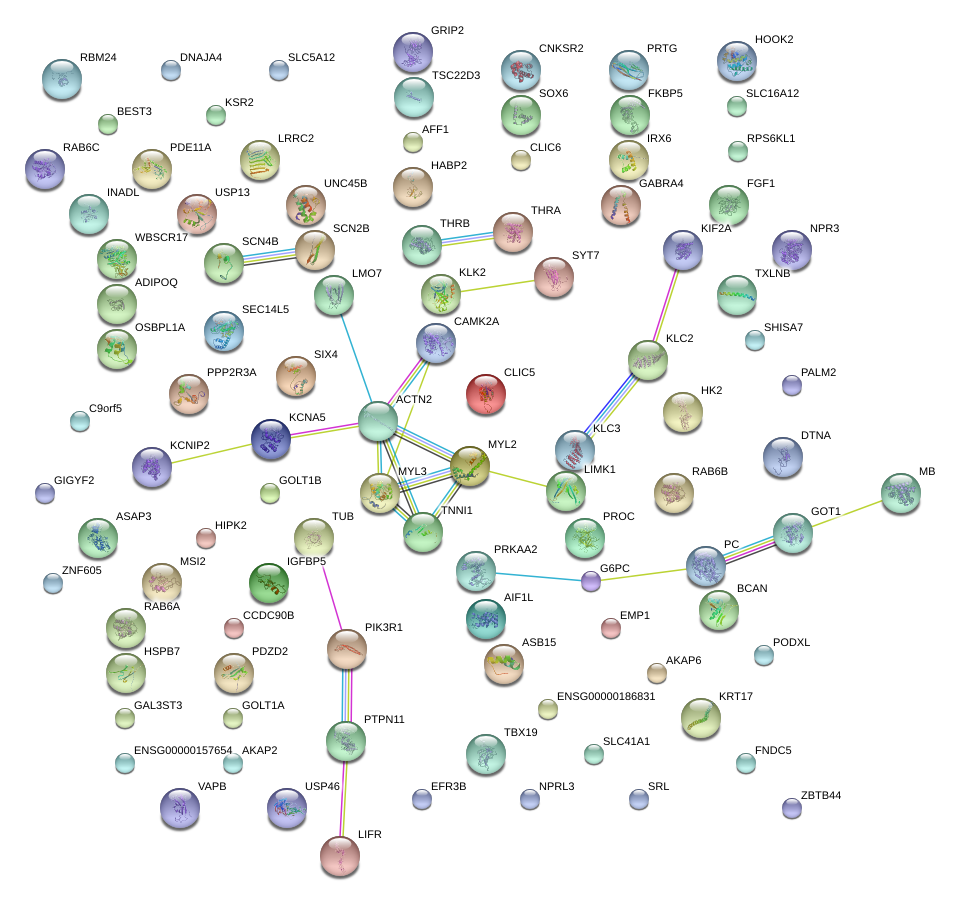
|
chr10_+_115312777
|
7.582
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr10_+_115310589
|
6.840
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr14_+_32798478
|
4.686
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr7_+_140774031
|
4.105
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr6_+_17281773
|
3.741
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr22_-_36013303
|
3.677
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr6_+_17282931
|
3.512
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_17282294
|
3.455
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr22_-_36019363
|
3.321
|
NM_203377
|
MB
|
myoglobin
|
|
chr5_-_142077508
|
3.158
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_141993691
|
3.073
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_142065991
|
2.986
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_142000899
|
2.899
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_-_142065586
|
2.666
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_33336289
|
2.110
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr1_-_33338081
|
2.069
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr1_-_16345284
|
1.890
|
NM_014424
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr12_-_70093064
|
1.783
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr17_+_55333843
|
1.782
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr1_+_62208091
|
1.605
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr7_-_139477437
|
1.579
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr12_-_111358353
|
1.536
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr4_-_53525316
|
1.499
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr11_-_61348290
|
1.475
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr4_+_87856152
|
1.474
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr6_-_46047978
|
1.468
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr12_-_70083055
|
1.439
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr3_-_46904879
|
1.429
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr6_-_139613114
|
1.414
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr1_+_236849753
|
1.356
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr3_+_135684463
|
1.328
|
NM_001190447
NM_002718
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr1_-_201390845
|
1.304
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr3_-_46608039
|
1.231
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr4_-_53522756
|
1.218
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr2_+_25264913
|
1.213
|
NM_014971
|
EFR3B
|
EFR3 homolog B (S. cerevisiae)
|
|
chr7_+_123249076
|
1.197
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr3_+_135741576
|
1.192
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr6_-_45983279
|
1.183
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr11_+_8060179
|
1.176
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr11_-_118016134
|
1.174
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr16_+_5008317
|
1.148
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr11_+_66025070
|
1.123
|
NM_001134776
NM_001134774
NM_022822
|
KLC2
|
kinesin light chain 2
|
|
chr17_+_33474835
|
1.121
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr9_+_133971862
|
1.118
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr5_+_32710742
|
1.101
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr2_-_178937472
|
1.088
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr11_-_118023534
|
1.074
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr11_+_66024746
|
1.056
|
NM_001134775
|
KLC2
|
kinesin light chain 2
|
|
chr2_-_178753464
|
1.048
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr14_-_61190458
|
1.042
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr5_-_38595506
|
1.032
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr11_-_118047336
|
0.991
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr11_-_66675334
|
0.950
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr11_-_26743545
|
0.949
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr5_+_31799030
|
0.918
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chrX_-_106960268
|
0.908
|
NM_004089
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr5_-_149669400
|
0.905
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr1_-_23810721
|
0.891
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr16_-_4292052
|
0.890
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr12_+_112856535
|
0.881
|
NM_002834
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr6_-_35696359
|
0.873
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr11_-_66725790
|
0.871
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr3_+_186560462
|
0.849
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr14_-_75389144
|
0.841
|
NM_031464
|
RPS6KL1
|
ribosomal protein S6 kinase-like 1
|
|
chr20_+_56964162
|
0.837
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr3_-_14583587
|
0.837
|
NM_001080423
|
GRIP2
|
glutamate receptor interacting protein 2
|
|
chr2_-_178973065
|
0.832
|
NM_001077197
|
PDE11A
|
phosphodiesterase 11A
|
|
chr2_-_217560247
|
0.822
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr9_+_112403067
|
0.819
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr3_-_24536176
|
0.810
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr11_-_16430425
|
0.790
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr4_+_87928083
|
0.790
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr5_+_32711437
|
0.789
|
NM_000908
NM_001204375
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr4_-_46995440
|
0.786
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr12_+_13349601
|
0.783
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr11_+_8102691
|
0.780
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr1_-_205782135
|
0.779
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr3_+_50242688
|
0.767
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr13_+_76194569
|
0.763
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr7_+_70597788
|
0.763
|
NM_022479
|
WBSCR17
|
Williams-Beuren syndrome chromosome region 17
|
|
chr10_-_103603539
|
0.762
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr12_-_118406027
|
0.739
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr1_+_26560613
|
0.731
|
NM_022778
|
CEP85
|
centrosomal protein 85kDa
|
|
chr1_+_57110745
|
0.730
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr11_-_130184459
|
0.730
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr18_+_32398225
|
0.725
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr17_+_55334373
|
0.717
|
NM_170721
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr18_+_32335939
|
0.713
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_+_75059781
|
0.710
|
NM_000189
|
HK2
|
hexokinase 2
|
|
chr3_+_179370779
|
0.710
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr7_+_73507485
|
0.708
|
NM_001204426
|
LIMK1
|
LIM domain kinase 1
|
|
chr4_-_46996423
|
0.704
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr11_-_16497917
|
0.699
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr10_+_99344079
|
0.690
|
NM_001134670
NM_138413
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr3_-_133614421
|
0.671
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr15_+_78556485
|
0.656
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_-_56035176
|
0.647
|
NM_173814
|
PRTG
|
protogenin
|
|
chr5_+_67588395
|
0.645
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr9_-_111882209
|
0.642
|
NM_032012
|
C9orf5
|
chromosome 9 open reading frame 5
|
|
chr11_-_65816650
|
0.629
|
NM_033036
|
GAL3ST3
|
galactose-3-O-sulfotransferase 3
|
|
chr10_-_101190201
|
0.626
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr2_+_233561997
|
0.623
|
NM_001103146
NM_001103147
NM_001103148
NM_015575
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr10_-_91295310
|
0.619
|
NM_213606
|
SLC16A12
|
solute carrier family 16, member 12 (monocarboxylic acid transporter 12)
|
|
chr16_+_55358470
|
0.610
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr14_+_60712469
|
0.602
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr17_+_5973640
|
0.602
|
NM_015253
|
WSCD1
|
WSC domain containing 1
|
|
chrX_-_142723921
|
0.596
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr2_-_213403280
|
0.596
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr15_+_78556901
|
0.595
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr10_-_103599596
|
0.595
|
NM_173194
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr15_-_90222572
|
0.580
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr18_-_48346297
|
0.577
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr4_-_47840058
|
0.576
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr13_+_76334796
|
0.574
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr6_+_118228688
|
0.574
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr7_+_73498106
|
0.573
|
NM_002314
|
LIMK1
|
LIM domain kinase 1
|
|
chr2_+_86333304
|
0.571
|
NM_017952
|
PTCD3
|
Pentatricopeptide repeat domain 3
|
|
chr1_-_216896640
|
0.568
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217262946
|
0.565
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr13_-_36050758
|
0.562
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr17_+_34058678
|
0.561
|
NM_033315
|
RASL10B
|
RAS-like, family 10, member B
|
|
chr1_+_53192021
|
0.558
|
NM_024646
|
ZYG11B
|
zyg-11 homolog B (C. elegans)
|
|
chr11_+_118478305
|
0.550
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr8_-_103251002
|
0.549
|
NM_001172477
NM_001172478
NM_015713
|
RRM2B
|
ribonucleotide reductase M2 B (TP53 inducible)
|
|
chr11_-_35441099
|
0.548
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_+_132129154
|
0.548
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr6_+_163835668
|
0.546
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr13_+_96743092
|
0.543
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr11_-_35441501
|
0.542
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr2_+_85198176
|
0.528
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr21_-_16437125
|
0.527
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chrX_-_111326003
|
0.526
|
NM_012471
|
TRPC5
|
transient receptor potential cation channel, subfamily C, member 5
|
|
chr10_-_116444374
|
0.526
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_-_144931967
|
0.515
|
NM_001002811
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr3_+_126707380
|
0.511
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr17_+_11144739
|
0.511
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr1_+_178310605
|
0.509
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr14_+_74318506
|
0.506
|
NM_001146154
NM_001146155
NM_152444
|
PTGR2
|
prostaglandin reductase 2
|
|
chr15_+_85923739
|
0.504
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr15_+_92937104
|
0.504
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr1_+_178062829
|
0.501
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr6_-_35656677
|
0.500
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr17_+_21279667
|
0.496
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr5_+_67511578
|
0.493
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr3_+_50712671
|
0.493
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr18_+_32397925
|
0.492
|
NM_001198943
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_+_30614815
|
0.491
|
NM_001109938
NM_001161376
NM_145029
|
C6orf136
|
chromosome 6 open reading frame 136
|
|
chr1_-_217311095
|
0.490
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_-_38556682
|
0.490
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr13_+_30002776
|
0.489
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr10_-_116418056
|
0.488
|
NM_002313
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_+_166326156
|
0.481
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr16_+_14164879
|
0.480
|
NM_014048
|
MKL2
|
MKL/myocardin-like 2
|
|
chr5_+_141488314
|
0.480
|
NM_030571
|
NDFIP1
|
Nedd4 family interacting protein 1
|
|
chr10_-_116286661
|
0.477
|
NM_006720
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr1_+_181452685
|
0.477
|
NM_000721
NM_001205293
NM_001205294
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr13_+_29598746
|
0.476
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr16_+_23568833
|
0.474
|
NM_019116
|
UBFD1
|
ubiquitin family domain containing 1
|
|
chr7_+_107301079
|
0.472
|
NM_000441
|
SLC26A4
|
solute carrier family 26, member 4
|
|
chr17_-_62050200
|
0.465
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr2_+_85980600
|
0.461
|
NM_032827
|
ATOH8
|
atonal homolog 8 (Drosophila)
|
|
chr16_+_70147528
|
0.455
|
NM_017990
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr1_+_109792640
|
0.454
|
NM_001408
|
CELSR2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr11_+_118477212
|
0.453
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr1_-_179112178
|
0.451
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr11_+_35684342
|
0.451
|
NM_017583
|
TRIM44
|
tripartite motif containing 44
|
|
chr9_+_87284573
|
0.450
|
NM_001018065
NM_001018066
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr1_-_154832187
|
0.449
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr15_-_23892992
|
0.446
|
NM_019066
|
MAGEL2
|
MAGE-like 2
|
|
chr15_-_40545109
|
0.445
|
NM_001039905
|
C15orf56
|
chromosome 15 open reading frame 56
|
|
chr3_+_101498028
|
0.444
|
NM_145037
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr11_+_107461807
|
0.443
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr16_-_19896105
|
0.443
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr18_-_59560303
|
0.441
|
NM_173557
|
RNF152
|
ring finger protein 152
|
|
chr7_-_73038814
|
0.439
|
NM_032951
NM_032952
NM_032953
NM_032954
|
MLXIPL
|
MLX interacting protein-like
|
|
chr2_+_103378489
|
0.436
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr7_+_55433140
|
0.436
|
NM_018697
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial)
|
|
chr2_-_44586854
|
0.428
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr14_-_88737160
|
0.424
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr20_+_48429249
|
0.424
|
NM_015266
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chrX_-_142722318
|
0.424
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr1_+_10270680
|
0.420
|
NM_015074
NM_183416
|
KIF1B
|
kinesin family member 1B
|
|
chr4_+_1902352
|
0.417
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr7_-_143599163
|
0.416
|
NM_014719
NM_001206938
|
FAM115A
|
family with sequence similarity 115, member A
|
|
chr3_+_101498285
|
0.416
|
NM_001134456
|
FAM55C
|
family with sequence similarity 55, member C
|
|
chr9_+_87284625
|
0.415
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr1_-_40157067
|
0.415
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr20_-_39928708
|
0.411
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr10_+_124768401
|
0.411
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr6_+_107811272
|
0.410
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chrX_+_77359665
|
0.410
|
NM_000291
|
PGK1
|
phosphoglycerate kinase 1
|
|
chr5_-_136834887
|
0.406
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr3_+_113465860
|
0.406
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr2_-_208890207
|
0.405
|
NM_001080475
|
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3
|
|
chr11_-_34379526
|
0.401
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr17_-_21454890
|
0.401
|
NM_001113434
|
C17orf51
|
chromosome 17 open reading frame 51
|
|
chr10_+_76871345
|
0.399
|
NM_001174156
NM_144660
|
SAMD8
|
sterile alpha motif domain containing 8
|
|
chr4_-_101111606
|
0.398
|
NM_145244
|
DDIT4L
|
DNA-damage-inducible transcript 4-like
|
|
chr2_-_44588622
|
0.397
|
NM_001171606
NM_001171613
|
PREPL
|
prolyl endopeptidase-like
|
|
chr10_+_121410751
|
0.393
|
NM_004281
|
BAG3
|
BCL2-associated athanogene 3
|