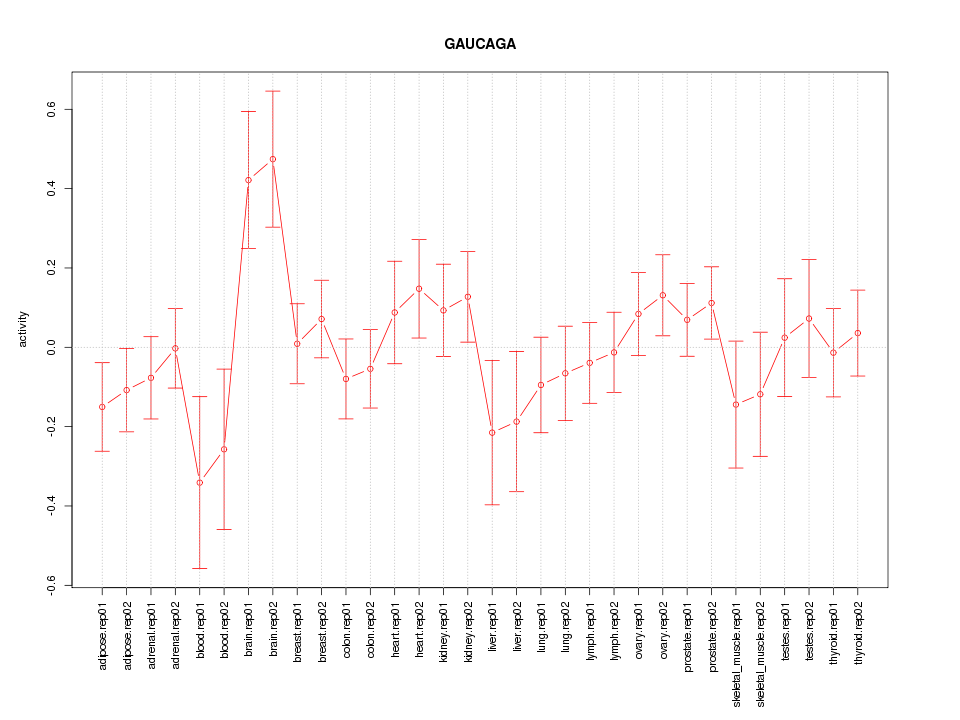
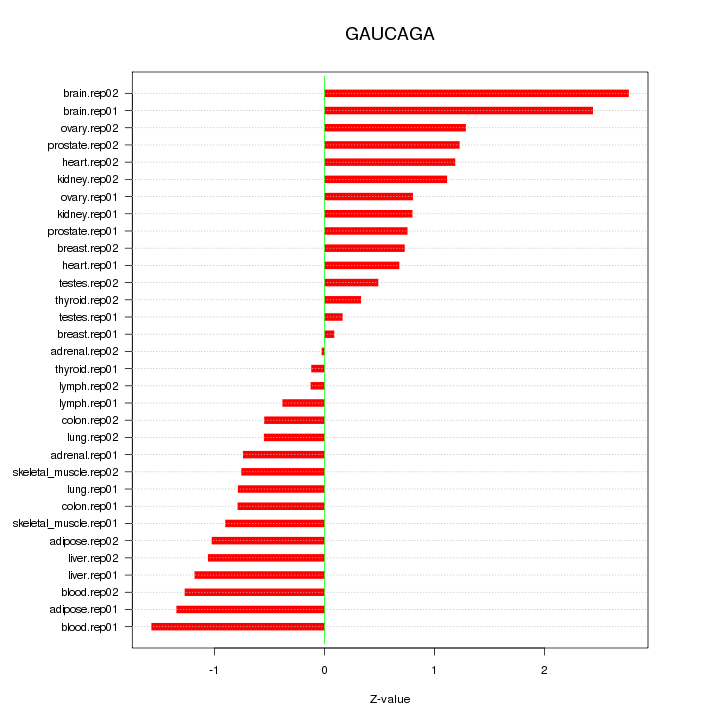
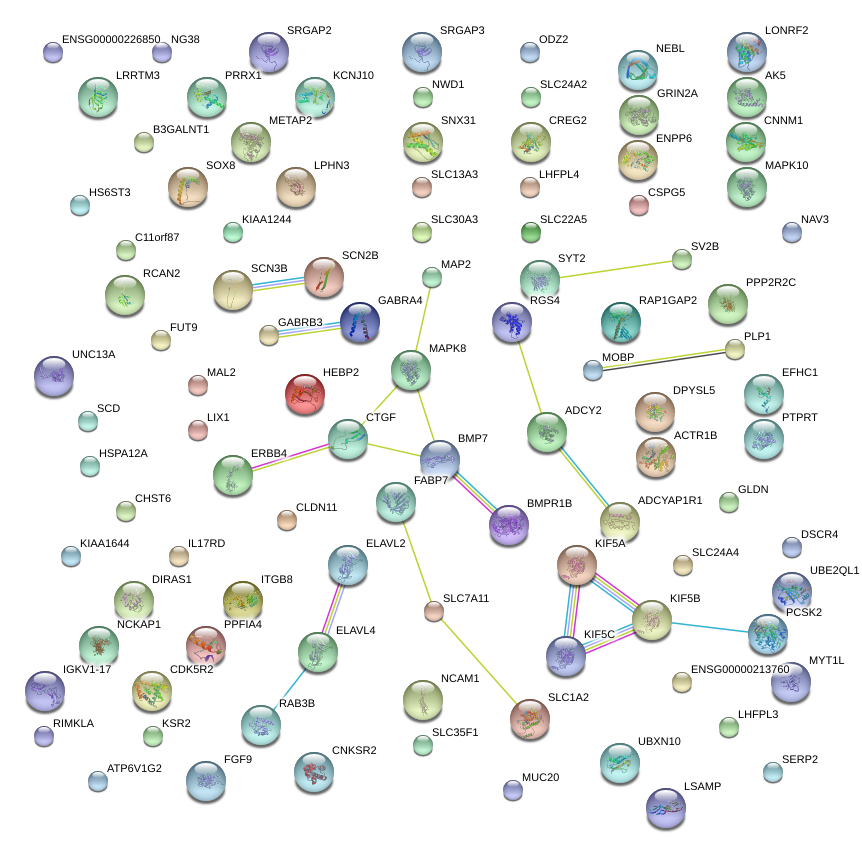
|
chr15_+_91643429
|
2.654
|
NM_014848
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr10_-_118501981
|
2.522
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr15_+_91643181
|
2.329
|
NM_001167580
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr16_-_10276610
|
2.231
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr15_+_51633712
|
2.152
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr1_-_52456292
|
2.133
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr16_-_10276115
|
2.128
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr12_-_118406027
|
1.891
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr8_+_120220554
|
1.774
|
NM_052886
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene)
|
|
chr16_-_75528925
|
1.559
|
NM_021615
|
CHST6
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6
|
|
chr19_+_16830786
|
1.516
|
NM_001007525
|
NWD1
|
NACHT and WD repeat domain containing 1
|
|
chr2_-_102003923
|
1.480
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr2_-_213403280
|
1.478
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr13_+_96743092
|
1.472
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr10_+_102106771
|
1.458
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr3_-_57199402
|
1.452
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr11_-_123525300
|
1.452
|
NM_001040151
NM_018400
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr3_-_9291251
|
1.391
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr3_+_195447751
|
1.388
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr1_-_160039946
|
1.373
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr1_+_163038934
|
1.365
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chrX_+_103031753
|
1.319
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr12_+_78225068
|
1.292
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr3_-_116164363
|
1.286
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr7_+_20370724
|
1.278
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr10_+_101088794
|
1.265
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chrX_+_103031433
|
1.251
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr6_-_31514620
|
1.247
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr5_+_7396030
|
1.199
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr1_+_20512577
|
1.195
|
NM_152376
|
UBXN10
|
UBX domain protein 10
|
|
chr17_+_2699731
|
1.185
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr19_-_2721337
|
1.182
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr6_+_96463844
|
1.166
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr4_-_185139018
|
1.164
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr13_+_22245083
|
1.145
|
NM_002010
|
FGF9
|
fibroblast growth factor 9 (glia-activating factor)
|
|
chr4_-_139163222
|
1.136
|
NM_014331
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11
|
|
chr2_-_100939194
|
1.131
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr4_-_6383572
|
1.125
|
NM_181876
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr1_+_163038564
|
1.082
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr4_-_6422844
|
1.072
|
NM_001206996
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr13_+_44947977
|
1.070
|
NM_001010897
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2
|
|
chr11_-_35441501
|
1.069
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_+_42846427
|
1.061
|
NM_173642
|
RIMKLA
|
ribosomal modification protein rimK-like family member A
|
|
chr3_+_195451097
|
1.058
|
NM_001098516
|
MUC20
|
mucin 20, cell surface associated
|
|
chr1_+_170633312
|
1.057
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr20_-_55841671
|
1.049
|
NM_001719
|
BMP7
|
bone morphogenetic protein 7
|
|
chr6_+_138483024
|
1.040
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr4_-_87374282
|
1.039
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_-_160822625
|
1.027
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr10_+_68685791
|
1.023
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr4_-_6557404
|
1.022
|
NM_001206995
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr4_-_46995440
|
1.022
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr11_-_118047336
|
1.012
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr4_-_6565326
|
1.004
|
NM_001206994
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr11_-_35441099
|
1.001
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_-_132272311
|
1.000
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr20_-_45280059
|
0.994
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr5_-_96478502
|
0.994
|
NM_153234
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr7_+_31092067
|
0.982
|
NM_001118
NM_001199635
NM_001199636
NM_001199637
|
ADCYAP1R1
|
adenylate cyclase activating polypeptide 1 (pituitary) receptor type I
|
|
chr5_+_166711842
|
0.973
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr9_-_23821477
|
0.967
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr20_-_45313122
|
0.964
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr2_-_183903225
|
0.953
|
NM_013436
NM_205842
|
NCKAP1
|
NCK-associated protein 1
|
|
chr9_-_23821842
|
0.949
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_2334887
|
0.941
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr20_+_17206751
|
0.940
|
NM_001201528
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_+_39509069
|
0.931
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr3_-_47620186
|
0.929
|
NM_001206943
NM_001206944
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr22_-_44708730
|
0.925
|
NM_001099294
|
KIAA1644
|
KIAA1644
|
|
chr4_+_95679075
|
0.923
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr6_+_118228688
|
0.922
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr2_+_149632772
|
0.921
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr20_-_41818464
|
0.919
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr3_+_170136652
|
0.915
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr11_+_109292837
|
0.914
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr6_+_52285804
|
0.912
|
NM_001172420
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr15_-_26874236
|
0.907
|
NM_001191321
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr10_-_21463019
|
0.906
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr9_-_19786886
|
0.904
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr22_-_36424397
|
0.902
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_203020310
|
0.892
|
NM_015053
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
|
|
chr5_+_6448735
|
0.892
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr6_-_46293628
|
0.891
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr1_+_163041694
|
0.889
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_-_202679415
|
0.888
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr2_+_27070962
|
0.880
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr19_-_17799005
|
0.872
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr2_-_27485774
|
0.871
|
NM_003459
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3
|
|
chr6_+_123100645
|
0.871
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr1_-_202612580
|
0.868
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr2_+_210288668
|
0.860
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr2_+_219824346
|
0.860
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr4_+_62362838
|
0.854
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr11_+_112831968
|
0.851
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr3_+_170139027
|
0.841
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr16_+_1031807
|
0.833
|
NM_014587
|
SOX8
|
SRY (sex determining region Y)-box 8
|
|
chr3_-_9595412
|
0.829
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr1_+_77748286
|
0.828
|
NM_012093
|
AK5
|
adenylate kinase 5
|
|
chr8_-_101661892
|
0.826
|
NM_152628
|
SNX31
|
sorting nexin 31
|
|
chr1_+_163038395
|
0.826
|
NM_001102445
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_-_111174095
|
0.824
|
NM_001204269
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2
|
|
chr8_+_79578258
|
0.817
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr10_+_43867089
|
0.815
|
NM_001184963
NM_173160
|
FXYD4
|
FXYD domain containing ion transport regulator 4
|
|
chr7_+_140774031
|
0.815
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr13_-_108519459
|
0.811
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr21_+_22370622
|
0.811
|
NM_004540
|
NCAM2
|
neural cell adhesion molecule 2
|
|
chr4_-_87028805
|
0.807
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_-_47621729
|
0.806
|
NM_001206942
NM_001206945
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr20_+_17207596
|
0.805
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr13_-_96296919
|
0.803
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr10_+_25464289
|
0.800
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr10_-_99393175
|
0.798
|
NM_178832
|
MORN4
|
MORN repeat containing 4
|
|
chr8_+_104512975
|
0.796
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr11_-_118016134
|
0.792
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr19_+_54466289
|
0.791
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr6_+_52284948
|
0.791
|
NM_018100
|
EFHC1
|
EF-hand domain (C-terminal) containing 1
|
|
chr5_-_148442606
|
0.791
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr21_-_48024985
|
0.785
|
NM_006272
|
S100B
|
S100 calcium binding protein B
|
|
chr15_-_26961911
|
0.781
|
NM_001191320
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr10_-_21186530
|
0.771
|
NM_006393
|
NEBL
|
nebulette
|
|
chr10_-_99393851
|
0.767
|
NM_001098831
|
MORN4
|
MORN repeat containing 4
|
|
chr11_+_61520000
|
0.760
|
NM_001127392
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr3_-_187388105
|
0.756
|
NM_001048
|
SST
|
somatostatin
|
|
chr4_-_96470350
|
0.754
|
NM_003728
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr14_+_85996454
|
0.751
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr13_-_45775174
|
0.750
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr1_+_151032150
|
0.745
|
NM_006818
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr10_-_93392857
|
0.741
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr2_-_217560247
|
0.741
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr7_-_79082870
|
0.739
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr10_-_118765087
|
0.727
|
NM_001127211
NM_018330
|
KIAA1598
|
KIAA1598
|
|
chr11_+_24518515
|
0.726
|
NM_001009909
NM_001252008
NM_001252010
|
LUZP2
|
leucine zipper protein 2
|
|
chr9_-_91793681
|
0.724
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr3_-_149688638
|
0.718
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chrX_+_70364680
|
0.717
|
NM_001166660
NM_018977
NM_181303
|
NLGN3
|
neuroligin 3
|
|
chr9_+_133971862
|
0.714
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr11_-_61348290
|
0.711
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr21_+_34398210
|
0.710
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr4_-_46996423
|
0.707
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr9_+_34458810
|
0.705
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr11_-_32452358
|
0.703
|
NM_001198551
NM_001198552
|
WT1
|
Wilms tumor 1
|
|
chr12_-_75723804
|
0.703
|
NM_032606
|
CAPS2
|
calcyphosine 2
|
|
chr11_-_118023534
|
0.703
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr11_+_61522860
|
0.698
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr11_-_83393436
|
0.695
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr2_+_210444364
|
0.694
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr4_-_87281374
|
0.688
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_+_110655061
|
0.686
|
NM_203412
|
UBL4B
|
ubiquitin-like 4B
|
|
chr17_+_48610045
|
0.685
|
NM_017957
|
EPN3
|
epsin 3
|
|
chr2_+_223916861
|
0.679
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr5_+_126626455
|
0.677
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr15_-_48470453
|
0.675
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr9_-_23826062
|
0.674
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_31845818
|
0.669
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr18_+_72201669
|
0.669
|
NM_032649
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family)
|
|
chr22_+_26565424
|
0.664
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr2_-_10220321
|
0.661
|
NM_001037160
|
CYS1
|
cystin 1
|
|
chr10_-_25241504
|
0.659
|
NM_020200
|
PRTFDC1
|
phosphoribosyl transferase domain containing 1
|
|
chr10_-_88126201
|
0.658
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr1_+_204913353
|
0.657
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr11_+_12399024
|
0.656
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr17_-_39890882
|
0.655
|
NM_001079870
NM_001079871
NM_177977
|
HAP1
|
huntingtin-associated protein 1
|
|
chr1_-_154832187
|
0.655
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr7_+_43152196
|
0.654
|
NM_015052
|
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1
|
|
chr11_+_107461807
|
0.653
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr6_-_132722564
|
0.652
|
NM_015529
|
MOXD1
|
monooxygenase, DBH-like 1
|
|
chr3_-_133614421
|
0.651
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr13_+_29598746
|
0.649
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr5_-_41510627
|
0.649
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr6_-_28411228
|
0.647
|
NM_001012455
|
ZSCAN23
|
zinc finger and SCAN domain containing 23
|
|
chr6_+_151561094
|
0.647
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr12_-_82153092
|
0.646
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr19_-_44324767
|
0.643
|
NM_182573
|
LYPD5
|
LY6/PLAUR domain containing 5
|
|
chr7_+_30174343
|
0.643
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr17_+_31254927
|
0.641
|
NM_001033504
NM_015544
|
TMEM98
|
transmembrane protein 98
|
|
chr4_-_53525316
|
0.639
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr8_-_143858436
|
0.634
|
NM_177476
NM_177477
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr11_-_84028379
|
0.634
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr8_-_133459508
|
0.634
|
NM_001204824
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr11_-_32457059
|
0.630
|
NM_000378
NM_024424
NM_024426
|
WT1
|
Wilms tumor 1
|
|
chr12_+_64238511
|
0.629
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr7_+_94285676
|
0.629
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr1_-_50489616
|
0.627
|
NM_032785
|
AGBL4
|
ATP/GTP binding protein-like 4
|
|
chr22_-_36236421
|
0.625
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_77747602
|
0.622
|
NM_174858
|
AK5
|
adenylate kinase 5
|
|
chr5_+_140552201
|
0.621
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr9_+_103947316
|
0.619
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr7_+_89840999
|
0.619
|
NM_001244944
NM_001244945
NM_152999
NM_001040665
NM_001244946
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr2_+_154728424
|
0.616
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr7_+_94285620
|
0.611
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr8_+_99439249
|
0.611
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr16_+_57406398
|
0.609
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr16_+_22825806
|
0.607
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr8_+_104831415
|
0.603
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr5_-_178017555
|
0.602
|
NM_173465
|
COL23A1
|
collagen, type XXIII, alpha 1
|
|
chr1_+_99729847
|
0.596
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr15_-_27018917
|
0.595
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr6_+_12290528
|
0.592
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr3_+_159706622
|
0.592
|
NM_000882
|
IL12A
|
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35)
|
|
chr12_-_81991959
|
0.590
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|