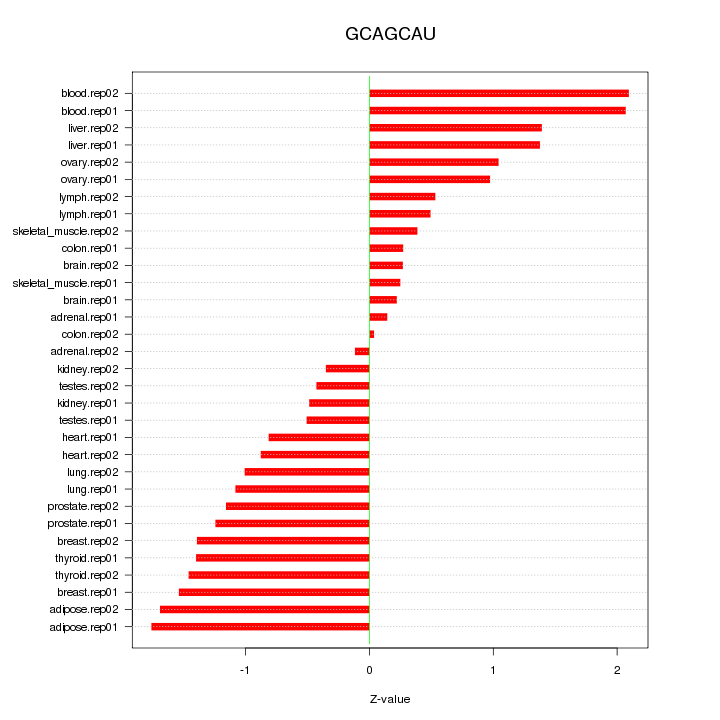
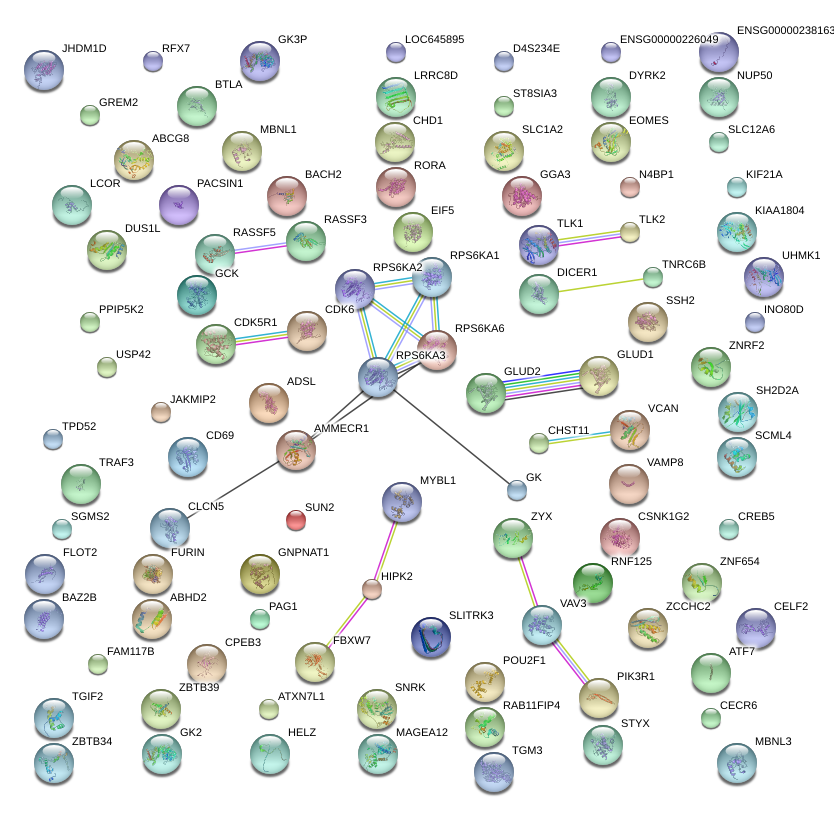
|
chr6_-_91006460
|
3.259
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr14_-_95599794
|
3.103
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr6_-_91006580
|
3.089
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_+_206680863
|
3.072
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr6_-_108145507
|
3.005
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr1_+_90287404
|
2.574
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chrX_+_49832214
|
2.477
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr3_-_27763784
|
2.352
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr1_+_90286469
|
2.321
|
NM_001134479
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr1_+_206730456
|
2.188
|
NM_182665
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr3_-_112218383
|
2.123
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr12_+_104850641
|
2.062
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr5_-_147162251
|
2.052
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr22_-_17602142
|
1.916
|
NM_031890
NM_001163079
|
CECR6
|
cat eye syndrome chromosome region, candidate 6
|
|
chr14_-_95608054
|
1.881
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr17_+_30813928
|
1.798
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chrX_+_30671456
|
1.731
|
NM_000167
NM_001128127
NM_001205019
NM_203391
|
GK
|
glycerol kinase
|
|
chr7_-_105319608
|
1.721
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr14_-_95623665
|
1.696
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chrX_-_131623994
|
1.685
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr3_-_164914448
|
1.649
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr5_+_67586466
|
1.646
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr8_-_82024277
|
1.646
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr5_+_82767492
|
1.619
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chrX_-_109561314
|
1.616
|
NM_001025580
NM_015365
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chrX_-_131573638
|
1.609
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr12_+_68042493
|
1.607
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr17_+_29718641
|
1.587
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr6_+_34433849
|
1.577
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr4_-_153273683
|
1.569
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_-_240775448
|
1.559
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr6_+_34482646
|
1.492
|
NM_001199583
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr4_+_4387982
|
1.485
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr22_+_40573879
|
1.474
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr10_-_94050799
|
1.469
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chrX_-_109683457
|
1.461
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr12_-_57400199
|
1.450
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chrX_-_131547536
|
1.418
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr18_+_29598444
|
1.356
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr1_-_156785938
|
1.349
|
NM_001161443
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr7_-_92465929
|
1.347
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr2_+_203499819
|
1.335
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr3_+_43328001
|
1.325
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr15_+_89631380
|
1.319
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chrX_-_20284708
|
1.311
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr7_-_92463211
|
1.300
|
NM_001259
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr4_+_108745718
|
1.271
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr22_-_39151457
|
1.256
|
NM_001199580
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr15_+_91411884
|
1.255
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr3_+_152017193
|
1.253
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr15_-_60884615
|
1.243
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr22_+_45560529
|
1.239
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr8_-_67525426
|
1.223
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr10_-_88854686
|
1.214
|
NM_005271
|
GLUD1
|
glutamate dehydrogenase 1
|
|
chr2_-_172087823
|
1.212
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr15_-_61521478
|
1.211
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chrX_+_49687212
|
1.208
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr7_+_143078358
|
1.192
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr7_+_6144504
|
1.187
|
NM_032172
|
USP42
|
ubiquitin specific peptidase 42
|
|
chr15_-_56535475
|
1.167
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr22_-_39152008
|
1.165
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr19_+_1941157
|
1.150
|
NM_001319
|
CSNK1G2
|
casein kinase 1, gamma 2
|
|
chr14_+_103243813
|
1.135
|
NM_001199427
NM_003300
NM_145725
NM_145726
|
TRAF3
|
TNF receptor-associated factor 3
|
|
chr22_+_40440664
|
1.130
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr12_-_39836760
|
1.126
|
NM_001173463
NM_001173464
NM_001173465
NM_017641
|
KIF21A
|
kinesin family member 21A
|
|
chr3_+_151985828
|
1.122
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr12_-_53994783
|
1.119
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr11_-_35441501
|
1.116
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr20_+_2276612
|
1.109
|
NM_003245
|
TGM3
|
transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr2_-_206950895
|
1.108
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr5_+_102465256
|
1.107
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr17_-_28257017
|
1.107
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr22_-_32146119
|
1.106
|
NM_173566
|
PRR14L
|
proline rich 14-like
|
|
chr14_+_53196852
|
1.099
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|
|
chr1_+_162466963
|
1.092
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr10_+_11206928
|
1.075
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_+_167298280
|
1.068
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_233463513
|
1.065
|
NM_032435
|
KIAA1804
|
mixed lineage kinase 4
|
|
chr4_-_153456152
|
1.063
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_-_108231122
|
1.060
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr2_-_171923482
|
1.057
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr3_+_88188261
|
1.038
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr7_-_105516923
|
1.036
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr2_+_44066102
|
1.033
|
NM_022437
|
ABCG8
|
ATP-binding cassette, sub-family G (WHITE), member 8
|
|
chr8_-_81083660
|
1.027
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr20_+_35201948
|
1.026
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr8_-_80993009
|
1.021
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr7_-_139477437
|
1.018
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr7_-_139876718
|
1.017
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr5_+_67588395
|
1.016
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr15_-_34629044
|
1.014
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_-_160472960
|
1.013
|
NM_013450
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr18_+_55019720
|
1.011
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr10_+_98592658
|
1.005
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr3_+_119187782
|
1.004
|
NM_152305
|
POGLUT1
|
protein O-glucosyltransferase 1
|
|
chr14_-_53258329
|
0.994
|
NM_198066
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1
|
|
chr1_-_156786500
|
0.989
|
NM_001161441
NM_001161442
NM_001161444
NM_003975
|
SH2D2A
|
SH2 domain containing 2A
|
|
chr12_-_9913415
|
0.988
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr10_-_94003002
|
0.980
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr10_+_11047253
|
0.966
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_-_98262217
|
0.961
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr5_+_67584251
|
0.959
|
NM_181524
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr11_-_35441099
|
0.954
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr17_-_65241305
|
0.951
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr20_+_35202956
|
0.947
|
NM_001199535
NM_001199515
|
TGIF2-C20ORF24
TGIF2
|
TGIF2-C20orf24 readthrough
TGFB-induced factor homeobox 2
|
|
chr2_+_85804613
|
0.935
|
NM_003761
|
VAMP8
|
vesicle-associated membrane protein 8 (endobrevin)
|
|
chr14_+_103801160
|
0.928
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr17_-_27224708
|
0.925
|
NM_004475
|
FLOT2
|
flotillin 2
|
|
chr7_+_30323922
|
0.924
|
NM_147128
|
ZNRF2
|
zinc and ring finger 2
|
|
chr1_-_108507474
|
0.916
|
NM_006113
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr18_+_60190657
|
0.913
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr17_-_73258441
|
0.909
|
NM_001172703
NM_001172704
|
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3
|
|
chr9_+_129622935
|
0.903
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr17_-_80023679
|
0.903
|
NM_022156
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae)
|
|
chr22_+_45559725
|
0.903
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr7_+_28475233
|
0.901
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_-_48644087
|
0.894
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr9_-_127905715
|
0.892
|
NM_001144877
NM_173690
|
SCAI
|
suppressor of cancer cell invasion
|
|
chr2_+_45878912
|
0.891
|
NM_005400
|
PRKCE
|
protein kinase C, epsilon
|
|
chr4_-_153303663
|
0.891
|
NM_001013415
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr1_-_173962050
|
0.889
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr1_+_26737232
|
0.887
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr1_-_220101925
|
0.886
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr14_+_57046490
|
0.884
|
NM_017799
|
C14orf101
|
chromosome 14 open reading frame 101
|
|
chr3_+_14989076
|
0.877
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chrX_-_131351917
|
0.876
|
NM_021183
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr20_+_35201854
|
0.866
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr10_+_92980322
|
0.865
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr8_+_32504250
|
0.864
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr4_-_78740508
|
0.862
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr12_+_57522247
|
0.853
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr2_-_16847070
|
0.839
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr17_+_26698986
|
0.838
|
NM_015077
|
SARM1
|
sterile alpha and TIR motif containing 1
|
|
chr22_-_42017032
|
0.837
|
NM_015704
|
PPPDE2
|
PPPDE peptidase domain containing 2
|
|
chr22_+_50781717
|
0.829
|
NM_001242898
NM_001242899
NM_001242900
NM_014678
|
PPP6R2
|
protein phosphatase 6, regulatory subunit 2
|
|
chr3_+_63884074
|
0.829
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr15_+_86220267
|
0.824
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr21_-_16437125
|
0.823
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr17_-_79479831
|
0.819
|
NM_001199954
NM_001614
|
ACTG1
|
actin, gamma 1
|
|
chr16_+_9185534
|
0.813
|
NM_014117
|
C16orf72
|
chromosome 16 open reading frame 72
|
|
chr12_-_49449106
|
0.809
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr7_+_28725597
|
0.807
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_-_34610928
|
0.805
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr5_-_111091947
|
0.804
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_39347562
|
0.802
|
NM_005633
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr12_-_57030083
|
0.801
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr17_-_27621163
|
0.789
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr4_+_3076236
|
0.785
|
NM_002111
|
HTT
|
huntingtin
|
|
chr10_+_11059854
|
0.785
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr2_-_219925237
|
0.784
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr15_-_34629941
|
0.779
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr4_+_57775064
|
0.770
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr12_+_105501448
|
0.768
|
NM_015275
|
KIAA1033
|
KIAA1033
|
|
chr8_-_57123858
|
0.766
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr16_+_50186809
|
0.764
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr7_+_28452143
|
0.761
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr1_-_151300168
|
0.761
|
NM_001198774
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta
|
|
chr4_+_174089889
|
0.760
|
NM_017423
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7)
|
|
chr1_+_36273786
|
0.760
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr9_-_139010716
|
0.760
|
NM_152833
|
C9orf69
|
chromosome 9 open reading frame 69
|
|
chr9_+_115513050
|
0.752
|
NM_001012994
|
SNX30
|
sorting nexin family member 30
|
|
chr15_-_34630203
|
0.747
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_-_201936389
|
0.743
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr12_-_46384365
|
0.741
|
NM_004719
|
SCAF11
|
SR-related CTD-associated factor 11
|
|
chr11_+_77300679
|
0.739
|
NM_173039
|
AQP11
|
aquaporin 11
|
|
chr3_-_13921617
|
0.738
|
NM_004625
|
WNT7A
|
wingless-type MMTV integration site family, member 7A
|
|
chr1_-_151300132
|
0.738
|
NM_001198773
NM_001198775
NM_002651
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta
|
|
chr12_+_104359557
|
0.737
|
NM_003211
|
TDG
|
thymine-DNA glycosylase
|
|
chr12_-_58240746
|
0.737
|
NM_005730
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr11_-_129062092
|
0.736
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr10_+_98592015
|
0.731
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr9_+_33817168
|
0.721
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr16_-_90039239
|
0.720
|
NM_145039
|
CENPBD1
|
CENPB DNA-binding domains containing 1
|
|
chr4_+_140222668
|
0.720
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr17_+_30264039
|
0.715
|
NM_015355
|
SUZ12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chrX_-_118827322
|
0.712
|
NM_015129
NM_145799
NM_145800
NM_145802
|
SEPT6
|
septin 6
|
|
chr2_+_208576239
|
0.708
|
NM_001142300
NM_152523
|
CCNYL1
|
cyclin Y-like 1
|
|
chr3_+_63953419
|
0.704
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr5_-_59783885
|
0.700
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr14_+_32546445
|
0.696
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr8_-_74791098
|
0.695
|
NM_001001481
NM_018299
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr17_+_61699786
|
0.692
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr1_+_162467594
|
0.692
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr11_-_16424391
|
0.685
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr2_-_198175494
|
0.684
|
NM_001195144
NM_153697
|
ANKRD44
|
ankyrin repeat domain 44
|
|
chr4_-_99579540
|
0.681
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr7_+_155250705
|
0.675
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr15_-_65579017
|
0.666
|
NM_017851
|
PARP16
|
poly (ADP-ribose) polymerase family, member 16
|
|
chr7_-_55640198
|
0.662
|
NM_030796
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1
|
|
chr20_+_1093905
|
0.661
|
NM_178578
|
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31)
|
|
chr7_-_121944485
|
0.661
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr5_+_5422792
|
0.654
|
NM_015325
|
KIAA0947
|
KIAA0947
|
|
chr1_+_36348795
|
0.654
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr18_-_3011944
|
0.644
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr16_-_70472920
|
0.643
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr2_-_46384
|
0.643
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr1_+_26438267
|
0.642
|
NM_001243533
NM_152835
|
PDIK1L
|
PDLIM1 interacting kinase 1 like
|
|
chr3_-_122233781
|
0.637
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr17_-_35969401
|
0.636
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr20_+_37434294
|
0.635
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|