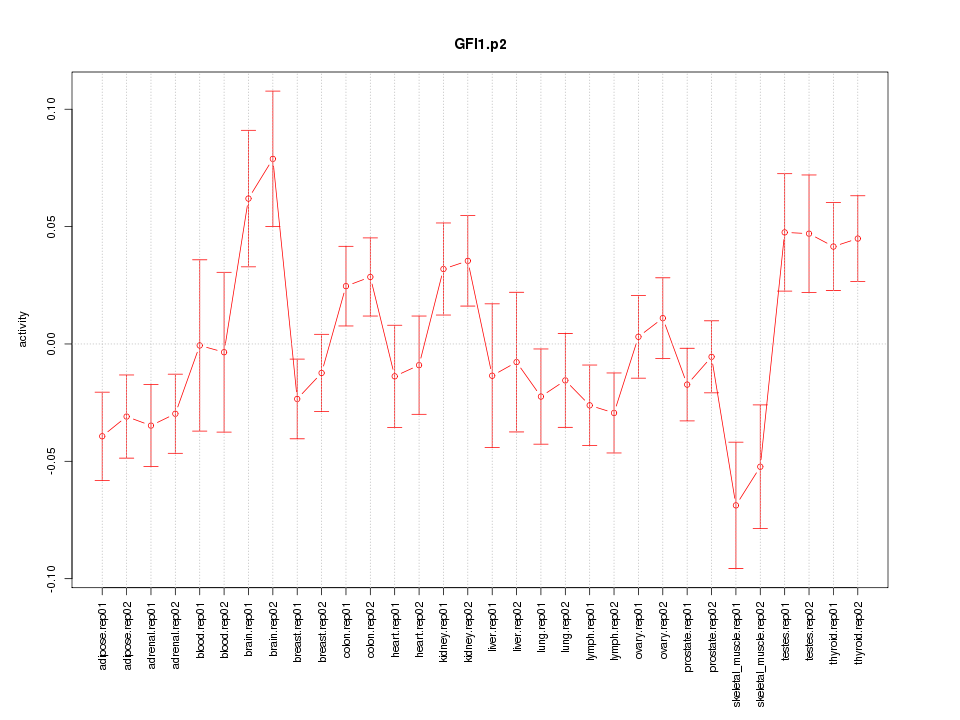
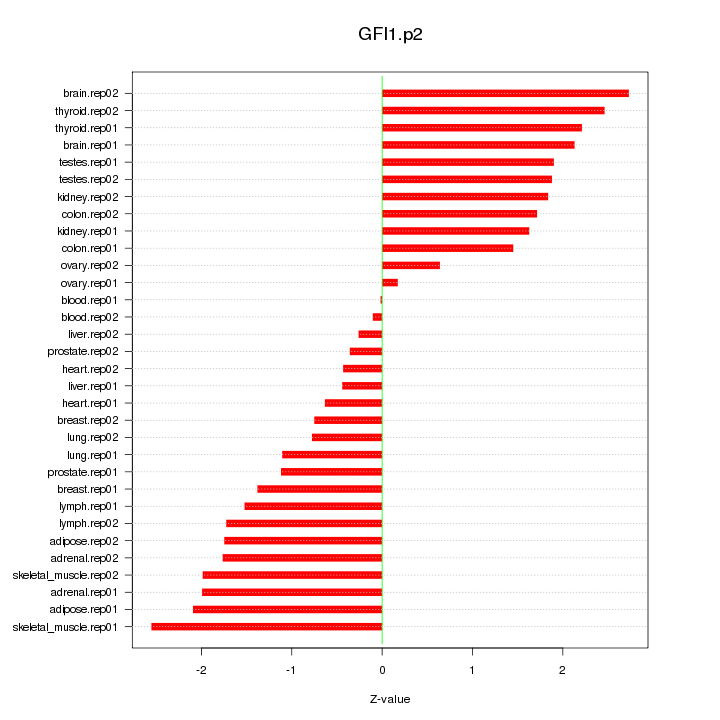
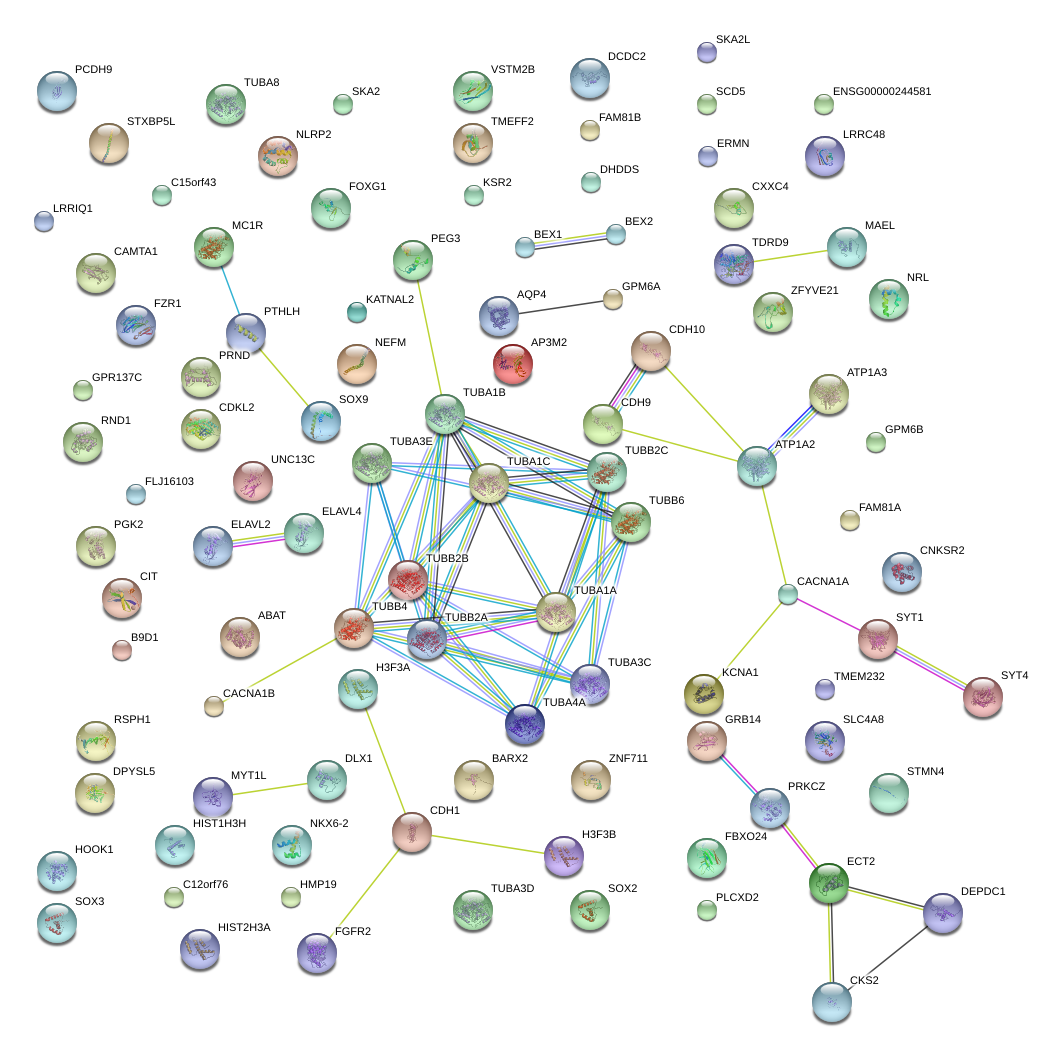
|
chr4_-_76555570
|
4.244
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr1_+_166958497
|
3.443
|
NM_032858
|
MAEL
|
maelstrom homolog (Drosophila)
|
|
chr21_-_43916298
|
3.343
|
NM_080860
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas)
|
|
chr14_+_104182045
|
2.643
|
NM_001198953
NM_024071
|
ZFYVE21
|
zinc finger, FYVE domain containing 21
|
|
chr12_-_120315074
|
2.612
|
NM_001206999
NM_007174
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr18_-_24445652
|
2.606
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chrX_-_102319096
|
2.583
|
NM_018476
|
BEX1
|
brain expressed, X-linked 1
|
|
chr17_+_70117168
|
2.534
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chrX_-_102319080
|
2.445
|
|
BEX1
|
brain expressed, X-linked 1
|
|
chr17_+_70117154
|
2.399
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr12_+_51818592
|
2.326
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr2_+_172950207
|
2.318
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr17_-_73775822
|
2.219
|
NM_005324
|
H3F3B
|
H3 histone, family 3B (H3.3B)
|
|
chr19_-_57352055
|
2.212
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr17_+_70117177
|
2.180
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr9_-_23826062
|
2.115
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_+_84498993
|
2.051
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr8_-_27115897
|
2.042
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr17_-_57232524
|
2.010
|
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr10_-_123357503
|
2.008
|
NM_000141
NM_001144917
NM_001144918
NM_001144919
NM_022970
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr8_+_42010463
|
2.000
|
NM_001134296
NM_006803
|
AP3M2
|
adaptor-related protein complex 3, mu 2 subunit
|
|
chr13_-_19755934
|
1.995
|
NM_006001
|
TUBA3C
|
tubulin, alpha 3c
|
|
chr12_+_79258588
|
1.951
|
|
SYT1
|
synaptotagmin I
|
|
chr15_+_59730272
|
1.915
|
NM_152450
|
FAM81A
|
family with sequence similarity 81, member A
|
|
chr12_+_85430098
|
1.903
|
NM_001079910
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr3_+_181429711
|
1.893
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr3_+_111393522
|
1.889
|
NM_001185106
NM_153268
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr14_+_53019865
|
1.827
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr2_-_165424993
|
1.827
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr5_-_27038650
|
1.823
|
NM_016279
|
CDH9
|
cadherin 9, type 2 (T1-cadherin)
|
|
chr18_+_44526786
|
1.814
|
NM_031303
|
KATNAL2
|
katanin p60 subunit A-like 2
|
|
chr5_+_173472692
|
1.807
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr17_-_57232799
|
1.777
|
NM_001100595
NM_182620
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr17_-_19281494
|
1.777
|
NM_001243475
|
B9D1
|
B9 protein domain 1
|
|
chr12_-_49259549
|
1.755
|
NM_014470
|
RND1
|
Rho family GTPase 1
|
|
chrX_+_84499037
|
1.729
|
|
ZNF711
|
zinc finger protein 711
|
|
chr1_-_38230804
|
1.676
|
NM_001099439
NM_173641
|
EPHA10
|
EPH receptor A10
|
|
chr12_+_51818669
|
1.672
|
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr9_+_91926109
|
1.668
|
NM_001827
|
CKS2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr19_+_36505409
|
1.657
|
|
|
|
|
chr12_-_118406027
|
1.628
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr2_-_193059599
|
1.621
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr7_+_100183926
|
1.619
|
NM_033506
|
FBXO24
|
F-box protein 24
|
|
chr5_+_173472737
|
1.614
|
|
HMP19
|
HMP19 protein
|
|
chr10_-_123356158
|
1.607
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr19_+_55477710
|
1.593
|
NM_001174082
NM_001174083
NM_017852
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chrX_-_139587224
|
1.583
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr3_+_172468469
|
1.575
|
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr3_+_119813600
|
1.565
|
|
|
|
|
chr9_+_140772228
|
1.563
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr4_-_105416050
|
1.537
|
|
CXXC4
|
CXXC finger protein 4
|
|
chr15_+_54305100
|
1.529
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr10_-_134599477
|
1.510
|
NM_177400
|
NKX6-2
|
NK6 homeobox 2
|
|
chr8_+_24771276
|
1.509
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr1_+_60280462
|
1.509
|
NM_015888
|
HOOK1
|
hook homolog 1 (Drosophila)
|
|
chr8_+_24771268
|
1.499
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr2_-_158182154
|
1.497
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr12_-_110505499
|
1.491
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr2_+_27070962
|
1.490
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr8_-_27115935
|
1.489
|
|
STMN4
|
stathmin-like 4
|
|
chr4_-_176734181
|
1.479
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chrX_-_13835012
|
1.475
|
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_1981902
|
1.475
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr3_+_120627026
|
1.460
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr10_-_123353330
|
1.446
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_-_193059249
|
1.443
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr14_+_104394751
|
1.427
|
NM_153046
|
TDRD9
|
tudor domain containing 9
|
|
chr15_+_45248902
|
1.427
|
NM_152448
|
C15orf43
|
chromosome 15 open reading frame 43
|
|
chr16_+_68771192
|
1.420
|
NM_004360
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr11_+_129245834
|
1.413
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr12_+_79258517
|
1.405
|
|
SYT1
|
synaptotagmin I
|
|
chr4_-_83719896
|
1.401
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr19_-_42498320
|
1.389
|
NM_152296
|
ATP1A3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr12_+_5019048
|
1.386
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr5_-_110062365
|
1.383
|
NM_001039763
|
TMEM232
|
transmembrane protein 232
|
|
chr2_-_2334887
|
1.375
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr5_+_94727047
|
1.356
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr16_+_8768403
|
1.348
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr5_-_24645084
|
1.344
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chr2_+_132233579
|
1.336
|
NM_080386
|
TUBA3D
|
tubulin, alpha 3d
|
|
chr12_-_122240827
|
1.320
|
|
|
|
|
chr14_+_29236171
|
1.307
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr6_-_49754899
|
1.302
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr20_+_4702499
|
1.296
|
NM_012409
|
PRND
|
prion protein 2 (dublet)
|
|
chr1_+_6845381
|
1.280
|
NM_001195563
NM_001242701
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr16_+_89989686
|
1.278
|
NM_006086
|
TUBB3
|
tubulin, beta 3 class III
|
|
chr6_-_24358279
|
1.267
|
NM_016356
|
DCDC2
|
doublecortin domain containing 2
|
|
chr13_-_67804432
|
1.263
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr18_-_40857245
|
1.262
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr19_+_30017490
|
1.254
|
NM_001146339
|
VSTM2B
|
V-set and transmembrane domain containing 2B
|
|
chr17_+_17876126
|
1.252
|
NM_001130090
NM_001130091
NM_001130092
NM_031294
|
LRRC48
|
leucine rich repeat containing 48
|
|
chr14_-_24584125
|
1.251
|
|
NRL
|
neural retina leucine zipper
|
|
chr1_-_68962679
|
1.234
|
|
DEPDC1
|
DEP domain containing 1
|
|
chr12_-_28124915
|
1.229
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chrX_+_102631267
|
1.226
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr7_-_100254048
|
1.225
|
NM_016188
|
ACTL6B
|
actin-like 6B
|
|
chr6_+_99282579
|
1.223
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr11_-_119293848
|
1.220
|
|
THY1
|
Thy-1 cell surface antigen
|
|
chr9_-_99801365
|
1.220
|
NM_001333
|
CTSL2
|
cathepsin L2
|
|
chr12_-_371994
|
1.217
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr15_+_84115979
|
1.214
|
NM_003027
|
SH3GL3
|
SH3-domain GRB2-like 3
|
|
chr13_+_37006628
|
1.213
|
|
CCNA1
|
cyclin A1
|
|
chr1_-_109825767
|
1.208
|
NM_001005290
NM_001032291
NM_032636
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chr5_+_68800003
|
1.204
|
NM_001205255
|
OCLN
|
occludin
|
|
chr4_+_155665150
|
1.202
|
NM_004744
|
LRAT
|
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase)
|
|
chr3_+_133494275
|
1.194
|
|
TF
|
transferrin
|
|
chr1_-_151762905
|
1.190
|
NM_001083963
NM_001083964
NM_001083965
NM_006862
|
TDRKH
|
tudor and KH domain containing
|
|
chr16_+_8806825
|
1.187
|
NM_000663
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr16_+_68771214
|
1.176
|
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr1_-_68962726
|
1.171
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr8_-_57359148
|
1.165
|
NM_001135690
|
PENK
|
proenkephalin
|
|
chr12_+_79258448
|
1.160
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr2_+_47630246
|
1.159
|
NM_000251
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr1_-_47779651
|
1.157
|
NM_001048166
NM_003035
|
STIL
|
SCL/TAL1 interrupting locus
|
|
chr1_+_50574601
|
1.156
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_-_19988293
|
1.150
|
NM_001167667
NM_004934
|
CDH18
|
cadherin 18, type 2
|
|
chr7_-_137531585
|
1.148
|
NM_004717
|
DGKI
|
diacylglycerol kinase, iota
|
|
chr8_+_1952875
|
1.144
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr8_+_27631902
|
1.141
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr12_-_113658826
|
1.131
|
NM_138451
|
IQCD
|
IQ motif containing D
|
|
chr1_+_120254418
|
1.126
|
NM_006623
|
PHGDH
|
phosphoglycerate dehydrogenase
|
|
chr19_+_1491000
|
1.107
|
NM_138393
|
REEP6
|
receptor accessory protein 6
|
|
chr11_-_85397215
|
1.105
|
NM_152723
|
CCDC89
|
coiled-coil domain containing 89
|
|
chrX_+_102631408
|
1.092
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr7_-_137028433
|
1.085
|
|
PTN
|
pleiotrophin
|
|
chr7_+_154002194
|
1.071
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr11_+_1411089
|
1.066
|
NM_003957
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr4_+_151503076
|
1.061
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr19_+_2867332
|
1.054
|
NM_024967
|
ZNF556
|
zinc finger protein 556
|
|
chr15_-_41522860
|
1.052
|
NM_152596
|
EXD1
|
exonuclease 3'-5' domain containing 1
|
|
chr1_-_109825683
|
1.051
|
|
PSRC1
|
proline/serine-rich coiled-coil 1
|
|
chrX_+_69664704
|
1.051
|
NM_021120
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr4_+_39184023
|
1.051
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chr9_-_91793375
|
1.050
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr2_-_114036487
|
1.050
|
NM_003466
NM_013952
NM_013953
NM_013992
|
PAX8
|
paired box 8
|
|
chr7_+_94285736
|
1.048
|
|
PEG10
|
paternally expressed 10
|
|
chr6_-_87804773
|
1.047
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr18_+_616671
|
1.044
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr14_-_58764805
|
1.037
|
|
FLJ31306
|
uncharacterized LOC379025
|
|
chr11_-_40315663
|
1.036
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr1_-_240775448
|
1.036
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr7_+_94285676
|
1.035
|
NM_001172437
NM_001172438
|
PEG10
|
paternally expressed 10
|
|
chr2_+_47630291
|
1.035
|
|
MSH2
|
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
|
|
chr2_-_2335080
|
1.028
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr6_+_123100860
|
1.028
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr1_+_31769847
|
1.025
|
|
ZCCHC17
|
zinc finger, CCHC domain containing 17
|
|
chr1_+_50575465
|
1.024
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_46125929
|
1.023
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr12_+_79258794
|
1.022
|
|
SYT1
|
synaptotagmin I
|
|
chr3_-_190580464
|
1.020
|
NM_001146686
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr17_+_63133501
|
1.017
|
|
RGS9
|
regulator of G-protein signaling 9
|
|
chr1_-_240775388
|
1.016
|
|
GREM2
|
gremlin 2
|
|
chr4_-_90759446
|
1.009
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr6_+_99282597
|
1.008
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr12_+_52056547
|
1.000
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr2_-_50574891
|
0.995
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr11_-_119294242
|
0.993
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr12_+_106994914
|
0.990
|
NM_001206691
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr3_+_181430075
|
0.987
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr6_-_25726754
|
0.986
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr11_-_62689011
|
0.984
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr7_+_137531690
|
0.983
|
|
|
|
|
chr11_-_84634437
|
0.971
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr14_-_74416711
|
0.964
|
|
FAM161B
|
family with sequence similarity 161, member B
|
|
chr1_-_109825745
|
0.964
|
|
|
|
|
chr18_-_3874688
|
0.964
|
NM_001242762
NM_001242763
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr3_+_170136652
|
0.953
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr11_+_1463674
|
0.950
|
|
BRSK2
|
BR serine/threonine kinase 2
|
|
chr5_+_140261792
|
0.950
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr12_-_6579754
|
0.949
|
NM_014231
NM_016830
NM_199245
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1)
|
|
chr11_+_71900601
|
0.947
|
NM_016724
NM_016725
|
FOLR1
|
folate receptor 1 (adult)
|
|
chr20_-_54967221
|
0.941
|
NM_003600
NM_198433
NM_198434
NM_198435
NM_198436
NM_198437
|
AURKA
|
aurora kinase A
|
|
chr1_+_85527992
|
0.937
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr11_-_2721223
|
0.937
|
|
KCNQ1OT1
|
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding)
|
|
chr2_-_152830448
|
0.931
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr5_-_157079371
|
0.927
|
NM_007017
NM_178424
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr14_+_65878579
|
0.926
|
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr5_-_59995938
|
0.919
|
NM_001145208
NM_018369
|
DEPDC1B
|
DEP domain containing 1B
|
|
chr20_-_58508668
|
0.919
|
|
SYCP2
|
synaptonemal complex protein 2
|
|
chrX_+_110366304
|
0.918
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr8_+_102504661
|
0.913
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr6_+_26020677
|
0.912
|
NM_003529
|
HIST1H3A
|
histone cluster 1, H3a
|
|
chr7_-_100253976
|
0.911
|
|
|
|
|
chr11_-_62688732
|
0.904
|
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr7_+_87257704
|
0.899
|
NM_001134405
NM_001134406
NM_138290
|
RUNDC3B
|
RUN domain containing 3B
|
|
chr5_-_73937248
|
0.896
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr12_+_106976684
|
0.894
|
NM_213594
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr3_-_9439122
|
0.885
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr4_+_95679075
|
0.885
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr3_-_197686837
|
0.883
|
NM_032263
|
IQCG
|
IQ motif containing G
|
|
chr3_+_131100514
|
0.882
|
NM_001171905
NM_001171906
NM_152395
|
NUDT16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr1_-_38512446
|
0.877
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr5_+_55033844
|
0.876
|
NM_001142549
NM_001166533
NM_024415
|
DDX4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
|
|
chr3_+_119421865
|
0.871
|
NM_033364
|
C3orf15
|
chromosome 3 open reading frame 15
|
|
chr11_-_84634219
|
0.870
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_+_98246595
|
0.867
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr11_+_94706780
|
0.866
|
NM_018039
|
KDM4D
|
lysine (K)-specific demethylase 4D
|
|
chr17_-_41977966
|
0.864
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chr6_+_123100645
|
0.864
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chrX_+_113818550
|
0.859
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|