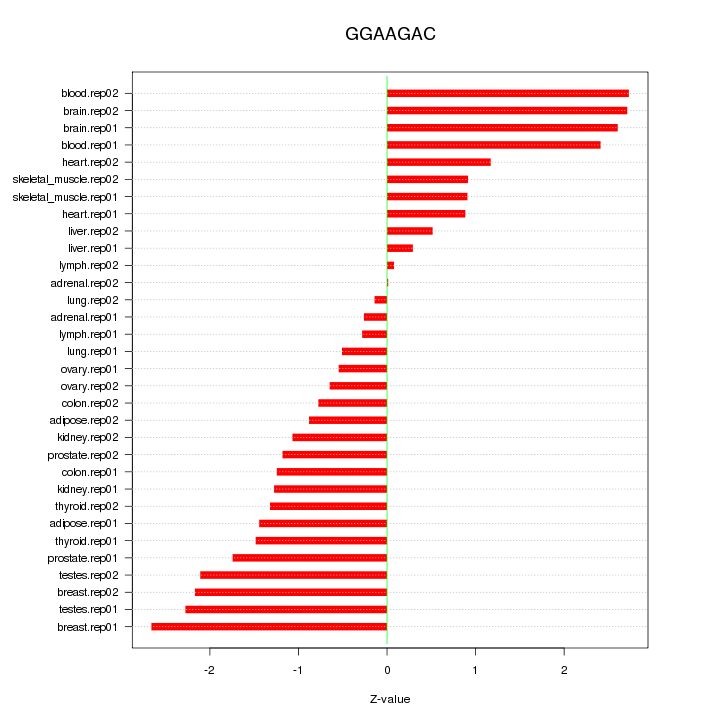
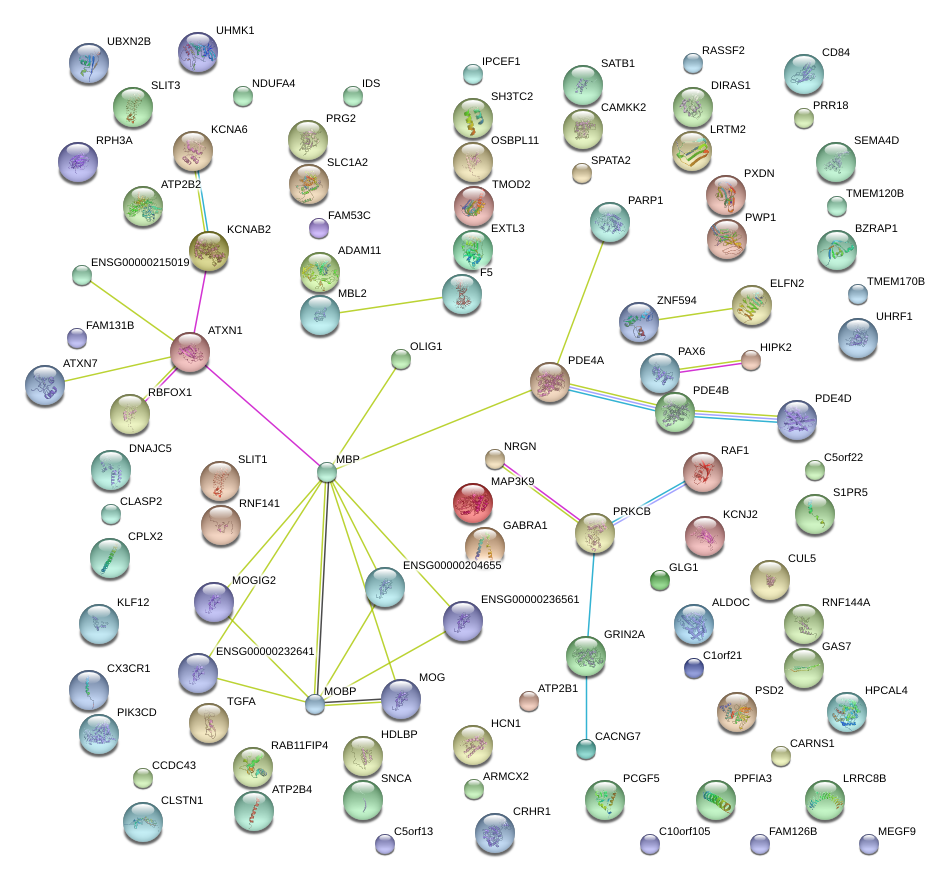
|
chr19_+_54415990
|
5.187
|
NM_031896
|
CACNG7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr17_+_68165660
|
4.005
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr5_+_161277702
|
3.633
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161275541
|
3.490
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr3_-_10547267
|
3.485
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr5_+_161274939
|
3.164
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161277391
|
3.138
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_90758126
|
2.974
|
NM_001146054
NM_007308
NM_000345
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chr10_-_98945662
|
2.971
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr5_+_161275897
|
2.961
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_162466963
|
2.950
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr5_+_161274196
|
2.908
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_9711781
|
2.898
|
NM_005026
|
PIK3CD
|
phosphoinositide-3-kinase, catalytic, delta polypeptide
|
|
chr5_+_161274684
|
2.891
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr17_+_29718641
|
2.761
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_-_90759446
|
2.725
|
NM_001146055
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor)
|
|
chrX_-_148586666
|
2.662
|
NM_000202
NM_001166550
NM_006123
|
IDS
|
iduronate 2-sulfatase
|
|
chr7_-_10979752
|
2.576
|
NM_002489
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr3_-_18480203
|
2.445
|
NM_001131010
|
SATB1
|
SATB homeobox 1
|
|
chr12_+_1929432
|
2.432
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr6_-_16761600
|
2.401
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr3_-_18466759
|
2.280
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr1_+_6105980
|
2.275
|
NM_001199862
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_-_35441501
|
2.245
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr3_-_33700932
|
2.216
|
NM_001207044
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr20_-_48530275
|
2.203
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr8_+_59323822
|
2.183
|
NM_001077619
|
UBXN2B
|
UBX domain protein 2B
|
|
chr5_+_31532372
|
2.175
|
NM_018356
|
C5orf22
|
chromosome 5 open reading frame 22
|
|
chr11_-_35441099
|
2.139
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr3_-_125314367
|
2.123
|
NM_022776
|
OSBPL11
|
oxysterol binding protein-like 11
|
|
chr5_-_148442606
|
2.114
|
NM_024577
|
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2
|
|
chr1_+_89990396
|
2.091
|
NM_001134476
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr11_-_31839447
|
2.040
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr3_-_39321479
|
1.979
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr1_+_6106173
|
1.958
|
NM_001199863
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr11_-_10562689
|
1.923
|
NM_016422
|
RNF141
|
ring finger protein 141
|
|
chr6_+_29624757
|
1.916
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr3_-_39323183
|
1.899
|
NM_001171174
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr16_+_23847271
|
1.883
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr17_-_56406120
|
1.817
|
NM_004758
NM_024418
|
BZRAP1
|
benzodiazapine receptor (peripheral) associated protein 1
|
|
chr1_+_184356128
|
1.815
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr5_-_45696219
|
1.802
|
NM_021072
|
HCN1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1
|
|
chr1_+_6086072
|
1.785
|
NM_003636
NM_172130
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr1_+_162467594
|
1.764
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr2_+_7057471
|
1.761
|
NM_014746
|
RNF144A
|
ring finger protein 144A
|
|
chr3_-_39322763
|
1.742
|
NM_001171171
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr1_+_90024271
|
1.740
|
NM_015350
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr1_+_6094342
|
1.737
|
NM_001199860
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr17_-_9929567
|
1.725
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr16_-_10276115
|
1.720
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr10_+_92980322
|
1.714
|
NM_032373
|
PCGF5
|
polycomb group ring finger 5
|
|
chr3_-_39321929
|
1.698
|
NM_001171172
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr16_-_10276610
|
1.674
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr5_-_58295758
|
1.672
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_63953419
|
1.667
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr3_-_33759684
|
1.662
|
NM_015097
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr5_-_58882274
|
1.635
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr20_+_62526453
|
1.633
|
NM_025219
|
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5
|
|
chr1_+_66797775
|
1.611
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_+_66258192
|
1.593
|
NM_001037341
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr13_-_74707913
|
1.586
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr14_-_71275732
|
1.586
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr6_+_11538486
|
1.582
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr1_+_66258855
|
1.569
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_+_6052357
|
1.539
|
NM_001199861
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr3_+_39509069
|
1.534
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr11_+_107879407
|
1.526
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr3_-_12705626
|
1.522
|
NM_002880
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr1_-_40157067
|
1.505
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr2_-_242212244
|
1.503
|
NM_001243900
NM_203346
|
HDLBP
|
high density lipoprotein binding protein
|
|
chr18_-_74729049
|
1.492
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr5_-_111092918
|
1.472
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr20_-_4804067
|
1.455
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr10_-_73479499
|
1.450
|
NM_001164375
|
C10orf105
|
chromosome 10 open reading frame 105
|
|
chr19_+_49622480
|
1.445
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr5_-_111093030
|
1.431
|
NM_001142482
NM_001142481
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_-_59783885
|
1.431
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_+_10563581
|
1.428
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr16_+_6823809
|
1.428
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr20_-_4795768
|
1.428
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr16_+_6069094
|
1.416
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_160549271
|
1.409
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr12_-_121734543
|
1.403
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr9_-_92094610
|
1.391
|
NM_001142287
NM_006378
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr17_+_42836546
|
1.382
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr5_-_111093251
|
1.379
|
NM_001142479
NM_001142480
NM_001142478
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr1_+_66458389
|
1.378
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr7_-_139477437
|
1.350
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr2_-_201936389
|
1.344
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr3_+_63884074
|
1.339
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr17_-_9940004
|
1.337
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr1_-_169555710
|
1.318
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr16_+_7382750
|
1.312
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr16_-_74641014
|
1.310
|
NM_001145666
NM_001145667
NM_012201
|
GLG1
|
golgi glycoprotein 1
|
|
chr17_-_42767016
|
1.310
|
NM_001099225
NM_144609
|
CCDC43
|
coiled-coil domain containing 43
|
|
chr19_+_10543110
|
1.293
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr17_-_9862765
|
1.284
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr6_-_154677868
|
1.264
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr7_-_143059144
|
1.258
|
NM_014690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr15_+_52043738
|
1.252
|
NM_001142885
NM_014548
|
TMOD2
|
tropomodulin 2 (neuronal)
|
|
chr12_+_113229548
|
1.238
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr11_+_124609775
|
1.217
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr6_-_154651214
|
1.206
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr10_-_73497580
|
1.201
|
NM_001168390
|
C10orf105
|
chromosome 10 open reading frame 105
|
|
chr13_+_115079943
|
1.182
|
NM_001164144
NM_001164145
NM_032436
|
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1
|
|
chr5_+_175223519
|
1.181
|
NM_006650
|
CPLX2
|
complexin 2
|
|
chr7_-_143059696
|
1.178
|
NM_001031690
|
FAM131B
|
family with sequence similarity 131, member B
|
|
chr11_-_31832855
|
1.176
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr19_-_10628111
|
1.174
|
NM_030760
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr17_-_10101854
|
1.160
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr19_-_2721337
|
1.142
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr8_+_28559143
|
1.133
|
NM_001440
|
EXTL3
|
exostoses (multiple)-like 3
|
|
chr21_+_34442446
|
1.128
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr12_+_122150638
|
1.127
|
NM_001080825
|
TMEM120B
|
transmembrane protein 120B
|
|
chr9_-_123476578
|
1.118
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr19_-_10628606
|
1.093
|
NM_001166215
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr1_-_226595796
|
1.092
|
NM_001618
|
PARP1
|
poly (ADP-ribose) polymerase 1
|
|
chr5_-_111312522
|
1.091
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr2_-_70781146
|
1.088
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr19_+_4910339
|
1.084
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr19_+_10531128
|
1.074
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr11_+_67183144
|
1.059
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr17_-_26903926
|
1.055
|
NM_005165
|
ALDOC
|
aldolase C, fructose-bisphosphate
|
|
chr5_+_139175385
|
1.053
|
NM_032289
|
PSD2
|
pleckstrin and Sec7 domain containing 2
|
|
chr12_+_4918341
|
1.045
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr22_-_37823503
|
1.040
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr6_-_166721870
|
1.038
|
NM_175922
|
PRR18
|
proline rich 18
|
|
chr5_-_111093792
|
1.036
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr5_+_137673702
|
1.029
|
NM_016605
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr5_+_175298492
|
1.029
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr17_-_29648631
|
1.010
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr12_+_129338051
|
0.999
|
NM_144669
|
GLT1D1
|
glycosyltransferase 1 domain containing 1
|
|
chr13_-_20806533
|
0.988
|
NM_001110219
NM_001110220
NM_001110221
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr17_-_77478562
|
0.984
|
NM_001082575
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr12_+_65563339
|
0.980
|
NM_001167614
NM_014319
|
LEMD3
|
LEM domain containing 3
|
|
chrX_-_19688991
|
0.978
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr5_-_131347249
|
0.976
|
NM_001009185
NM_001205247
NM_015256
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr18_+_9137560
|
0.967
|
NM_001204056
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr1_-_47069936
|
0.955
|
NM_001135553
NM_003684
NM_198973
|
MKNK1
|
MAP kinase interacting serine/threonine kinase 1
|
|
chr5_-_157002739
|
0.954
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr1_+_24742244
|
0.950
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr11_-_64646053
|
0.938
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr1_+_181452685
|
0.918
|
NM_000721
NM_001205293
NM_001205294
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr16_-_755718
|
0.917
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr5_-_65017874
|
0.903
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr17_-_28257017
|
0.898
|
NM_033389
|
SSH2
|
slingshot homolog 2 (Drosophila)
|
|
chr18_+_9136742
|
0.894
|
NM_001083625
NM_015208
|
ANKRD12
|
ankyrin repeat domain 12
|
|
chr8_+_26371461
|
0.887
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr20_+_57766074
|
0.880
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr1_-_160039946
|
0.879
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr3_-_195635879
|
0.870
|
NM_005781
|
TNK2
|
tyrosine kinase, non-receptor, 2
|
|
chr11_-_6640661
|
0.866
|
NM_000391
|
TPP1
|
tripeptidyl peptidase I
|
|
chr5_-_131347518
|
0.864
|
NM_001205248
NM_001205251
NM_001205250
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr11_-_59436488
|
0.863
|
NM_152716
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr15_+_64791618
|
0.856
|
NM_015042
|
ZNF609
|
zinc finger protein 609
|
|
chr14_-_77608132
|
0.853
|
NM_174976
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr3_-_151047309
|
0.851
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr1_-_182360400
|
0.848
|
NM_001033056
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_-_182361311
|
0.848
|
NM_001033044
NM_002065
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_-_194119994
|
0.844
|
NM_004488
|
GP5
|
glycoprotein V (platelet)
|
|
chr3_+_63850181
|
0.843
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr1_-_161601752
|
0.842
|
NM_000570
|
FCGR3B
|
Fc fragment of IgG, low affinity IIIb, receptor (CD16b)
|
|
chr5_+_175084984
|
0.839
|
NM_001131055
|
HRH2
|
histamine receptor H2
|
|
chr7_+_50344255
|
0.839
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr17_-_4167028
|
0.828
|
NM_016376
NM_020740
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr16_-_12010518
|
0.828
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr1_-_160832466
|
0.827
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr20_+_1875382
|
0.824
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr8_-_145018057
|
0.815
|
NM_201382
|
PLEC
|
plectin
|
|
chr1_-_161520412
|
0.815
|
NM_001127593
NM_001127595
NM_001127596
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr3_+_111260925
|
0.814
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr20_-_30310597
|
0.814
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr8_-_145016691
|
0.805
|
NM_201383
|
PLEC
|
plectin
|
|
chr5_-_59189620
|
0.802
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_+_6887576
|
0.802
|
NM_001974
|
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1
|
|
chr19_+_48111437
|
0.795
|
NM_015711
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1
|
|
chr18_-_3874252
|
0.793
|
NM_001242764
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr8_+_27183062
|
0.790
|
NM_173175
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr11_+_45868668
|
0.786
|
NM_001127457
|
CRY2
|
cryptochrome 2 (photolyase-like)
|
|
chr12_-_54694717
|
0.783
|
NM_001136023
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr1_-_162381805
|
0.781
|
NM_053282
|
SH2D1B
|
SH2 domain containing 1B
|
|
chr18_-_32924417
|
0.778
|
NM_006965
|
ZNF24
|
zinc finger protein 24
|
|
chr1_-_161519817
|
0.775
|
NM_000569
NM_001127592
|
FCGR3A
|
Fc fragment of IgG, low affinity IIIa, receptor (CD16a)
|
|
chr8_+_27179918
|
0.774
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr1_+_154377668
|
0.771
|
NM_000565
NM_001206866
NM_181359
|
IL6R
|
interleukin 6 receptor
|
|
chr1_+_151584514
|
0.770
|
NM_030918
|
SNX27
|
sorting nexin family member 27
|
|
chr3_-_172428773
|
0.768
|
NM_001146276
NM_001146277
NM_001146278
NM_020792
|
NCEH1
|
neutral cholesterol ester hydrolase 1
|
|
chr21_-_32931289
|
0.768
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr2_+_97426571
|
0.764
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr12_-_54689536
|
0.764
|
NM_006163
|
NFE2
|
nuclear factor (erythroid-derived 2), 45kDa
|
|
chr8_+_27168953
|
0.760
|
NM_173174
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr20_+_35234077
|
0.759
|
NM_001199534
NM_018840
NM_199483
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr5_+_132083135
|
0.755
|
NM_001039780
|
CCNI2
|
cyclin I family, member 2
|
|
chr1_-_217262946
|
0.750
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_-_111091947
|
0.749
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr19_-_17414083
|
0.747
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr1_-_109584589
|
0.747
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr8_-_145013628
|
0.742
|
NM_201384
|
PLEC
|
plectin
|
|
chr2_-_87018802
|
0.739
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr20_+_35202956
|
0.737
|
NM_001199535
NM_001199515
|
TGIF2-C20ORF24
TGIF2
|
TGIF2-C20orf24 readthrough
TGFB-induced factor homeobox 2
|