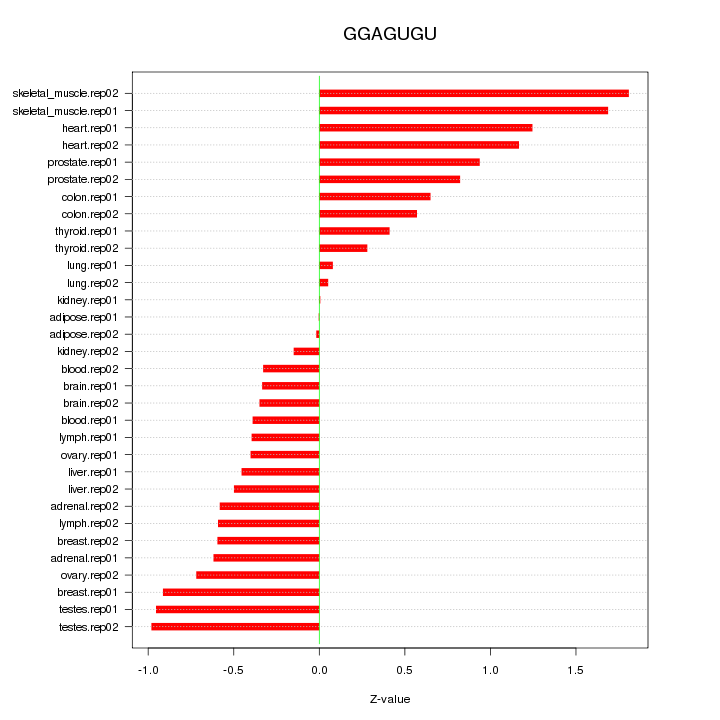
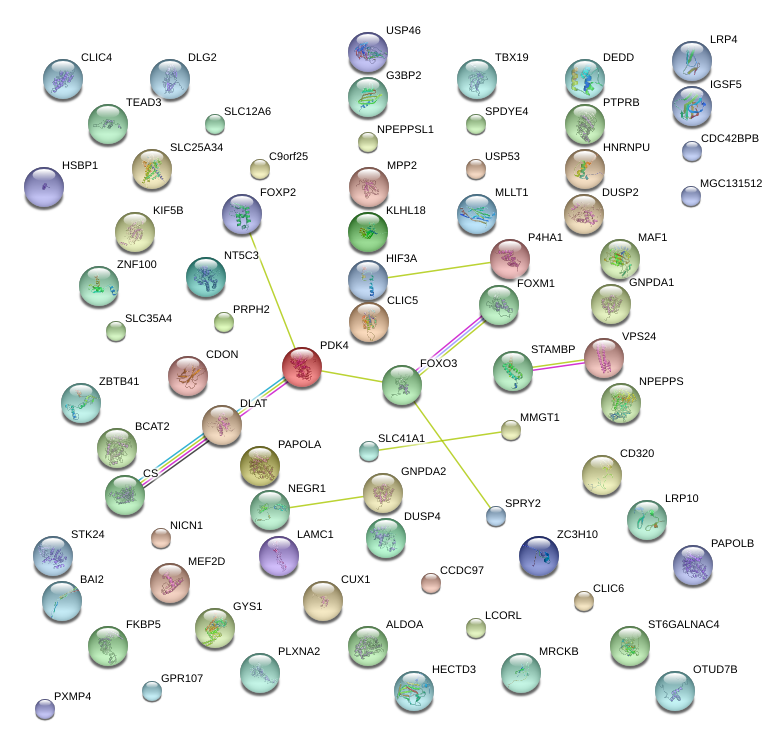
|
chr7_-_95225802
|
2.664
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr16_+_30064410
|
2.275
|
NM_000034
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr1_+_16062808
|
2.205
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr6_-_35696359
|
2.036
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr16_+_30075530
|
1.951
|
NM_184043
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr10_-_74856656
|
1.925
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|
|
chr19_-_49496566
|
1.867
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr16_+_30075715
|
1.745
|
NM_001127617
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr17_+_45608443
|
1.606
|
NM_006310
|
NPEPPS
|
aminopeptidase puromycin sensitive
|
|
chr6_-_35464716
|
1.557
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr21_+_41117333
|
1.269
|
NM_001080444
|
IGSF5
|
immunoglobulin superfamily, member 5
|
|
chr16_+_30076993
|
1.171
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr13_-_80915041
|
1.165
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr6_-_35656677
|
1.096
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr6_-_46047978
|
1.057
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr6_-_45983279
|
1.050
|
NM_016929
|
CLIC5
|
chloride intracellular channel 5
|
|
chr8_-_29206321
|
1.039
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr8_+_145159304
|
1.000
|
NM_032272
|
MAF1
|
MAF1 homolog (S. cerevisiae)
|
|
chr19_+_46800296
|
0.987
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr7_-_33080517
|
0.965
|
NM_001166118
NM_016489
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr11_+_111895537
|
0.956
|
NM_001931
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr12_-_56694174
|
0.946
|
NM_004077
|
CS
|
citrate synthase
|
|
chr7_+_101460881
|
0.942
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr1_-_156470506
|
0.892
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr19_+_46806855
|
0.815
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr2_-_86948244
|
0.759
|
NM_001198954
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough
|
|
chr2_-_86790533
|
0.744
|
NM_001005753
NM_001193517
NM_016079
|
CHMP3
|
charged multivesicular body protein 3
|
|
chr6_+_108882068
|
0.727
|
NM_001455
|
FOXO3
|
forkhead box O3
|
|
chr1_-_149982528
|
0.723
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr4_+_120133764
|
0.715
|
NM_019050
|
USP53
|
ubiquitin specific peptidase 53
|
|
chr6_+_108881010
|
0.710
|
NM_201559
|
FOXO3
|
forkhead box O3
|
|
chr19_+_46801632
|
0.676
|
NM_022462
NM_152796
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr7_-_33102174
|
0.675
|
NM_001002010
|
NT5C3
|
5'-nucleotidase, cytosolic III
|
|
chr7_+_114055048
|
0.650
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr1_-_205782135
|
0.632
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr8_-_29207795
|
0.620
|
NM_001394
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr12_+_56511969
|
0.606
|
NM_032786
|
ZC3H10
|
zinc finger CCCH-type containing 10
|
|
chr19_-_49314177
|
0.558
|
NM_001164773
NM_001190
|
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial
|
|
chr19_-_8373157
|
0.544
|
NM_001165895
NM_016579
|
CD320
|
CD320 molecule
|
|
chr4_-_53522756
|
0.528
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr9_-_34458477
|
0.453
|
NM_001184940
NM_001184941
NM_001184942
NM_001184943
NM_001184945
NM_147202
|
C9orf25
|
chromosome 9 open reading frame 25
|
|
chr1_-_208417635
|
0.428
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr5_-_141392537
|
0.377
|
NM_005471
|
GNPDA1
|
glucosamine-6-phosphate deaminase 1
|
|
chr1_+_25071759
|
0.372
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr4_-_53525316
|
0.347
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr4_-_76598289
|
0.333
|
NM_012297
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr1_-_72748147
|
0.318
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr11_-_85338313
|
0.297
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_-_71003622
|
0.295
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr3_+_47324329
|
0.282
|
NM_025010
|
KLHL18
|
kelch-like 18 (Drosophila)
|
|
chr1_-_32229635
|
0.266
|
NM_001703
|
BAI2
|
brain-specific angiogenesis inhibitor 2
|
|
chr9_+_132815984
|
0.253
|
NM_001136557
NM_001136558
NM_020960
|
GPR107
|
G protein-coupled receptor 107
|
|
chr14_+_23340955
|
0.244
|
NM_014045
|
LRP10
|
low density lipoprotein receptor-related protein 10
|
|
chr1_-_161102210
|
0.243
|
NM_001039712
|
DEDD
|
death effector domain containing
|
|
chr19_-_6279927
|
0.237
|
NM_005934
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1
|
|
chrX_-_135056132
|
0.222
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr4_-_76598611
|
0.218
|
NM_203504
NM_203505
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2
|
|
chr12_-_71031174
|
0.207
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr1_+_182992329
|
0.196
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr10_-_32345337
|
0.186
|
NM_004521
|
KIF5B
|
kinesin family member 5B
|
|
chr4_-_44728518
|
0.169
|
NM_138335
|
GNPDA2
|
glucosamine-6-phosphate deaminase 2
|
|
chr19_+_41816032
|
0.164
|
NM_052848
|
CCDC97
|
coiled-coil domain containing 97
|
|
chr15_-_34629044
|
0.158
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr15_-_34610928
|
0.155
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr1_-_45477016
|
0.136
|
NM_024602
|
HECTD3
|
HECT domain containing 3
|
|
chr1_-_161102457
|
0.134
|
NM_001039711
NM_032998
|
DEDD
|
death effector domain containing
|
|
chr1_-_197169635
|
0.130
|
NM_194314
|
ZBTB41
|
zinc finger and BTB domain containing 41
|
|
chr14_+_96968698
|
0.111
|
NM_001252006
NM_001252007
NM_032632
|
PAPOLA
|
poly(A) polymerase alpha
|
|
chr20_-_32307889
|
0.108
|
NM_007238
NM_183397
|
PXMP4
|
peroxisomal membrane protein 4, 24kDa
|
|
chr2_+_74056095
|
0.089
|
NM_006463
NM_213622
|
STAMBP
|
STAM binding protein
|
|
chr1_-_245027808
|
0.087
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr4_-_18023358
|
0.079
|
NM_001166139
NM_153686
|
LCORL
|
ligand dependent nuclear receptor corepressor-like
|
|
chr2_+_74056042
|
0.061
|
NM_201647
|
STAMBP
|
STAM binding protein
|
|
chr5_+_139944411
|
0.061
|
NM_080670
|
SLC35A4
|
solute carrier family 35, member A4
|
|
chr2_-_96811138
|
0.059
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr9_-_130679210
|
0.043
|
NM_175039
NM_175040
|
ST6GALNAC4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr3_-_49466744
|
0.039
|
NM_032316
|
NICN1
|
nicolin 1
|
|
chr13_-_99174251
|
0.034
|
NM_003576
|
STK24
|
serine/threonine kinase 24
|
|
chr13_-_99229226
|
0.032
|
NM_001032296
|
STK24
|
serine/threonine kinase 24
|
|
chr14_-_103523619
|
0.020
|
NM_006035
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like)
|