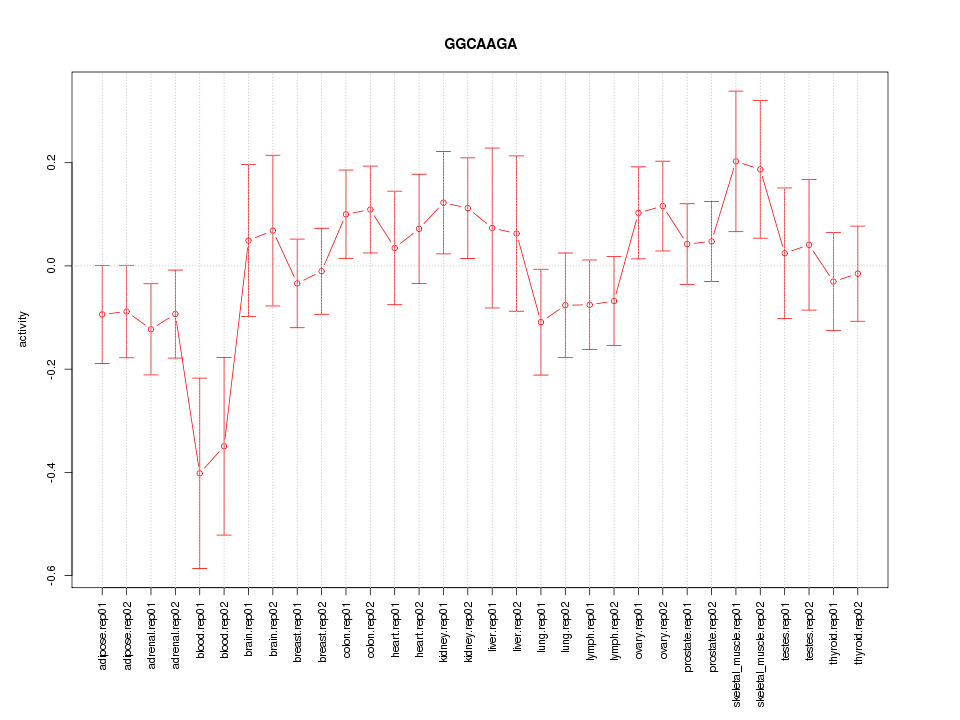
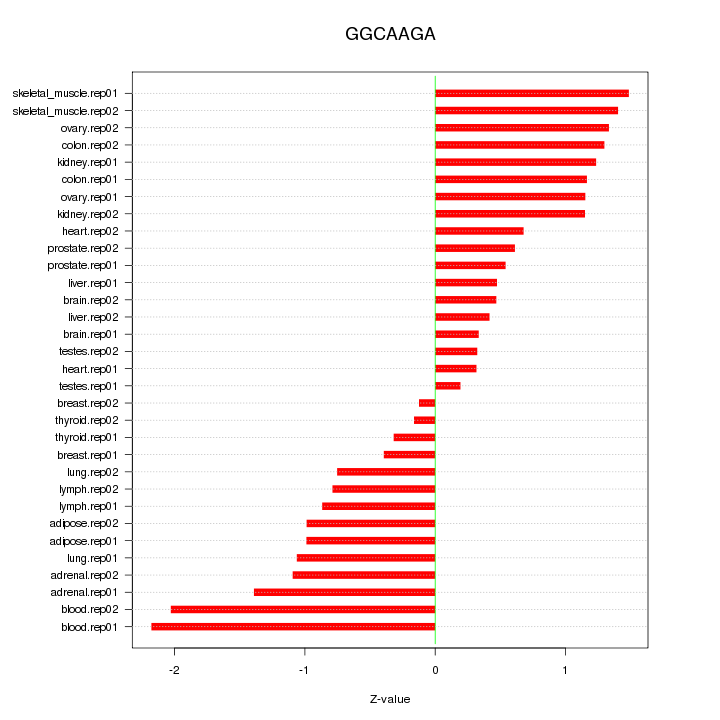
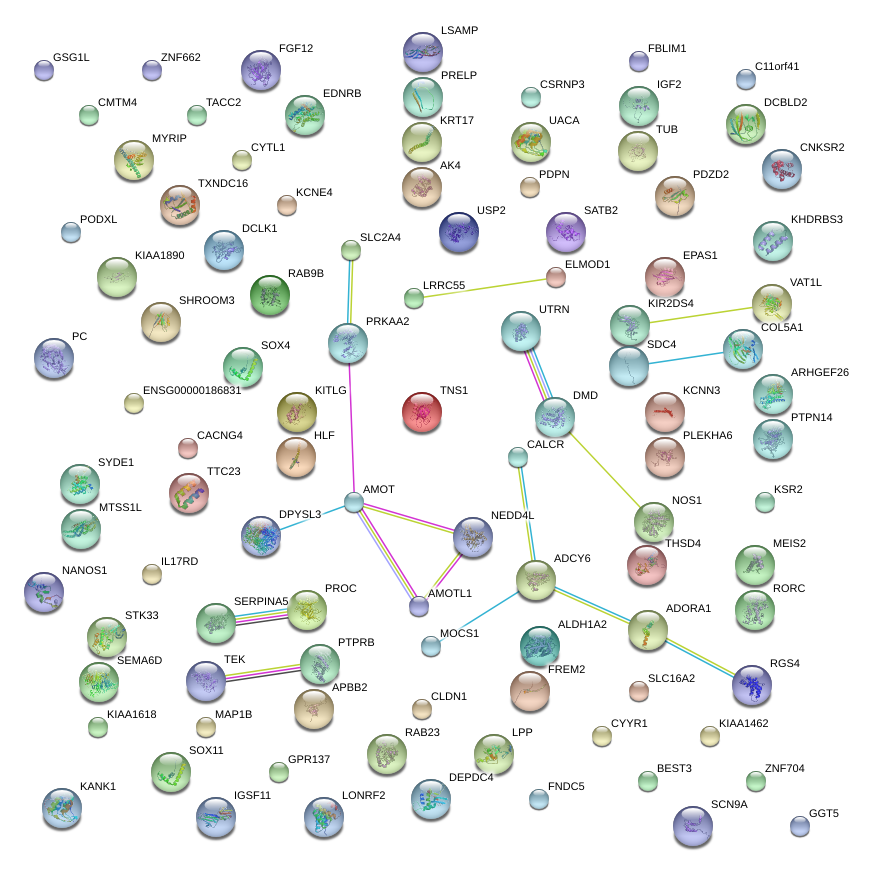
|
chr1_-_214724974
|
3.599
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr2_-_218808705
|
2.965
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr1_-_33336289
|
2.817
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr1_-_33338081
|
2.731
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr3_-_118864881
|
2.505
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr8_+_136469557
|
2.332
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr15_-_71055745
|
2.326
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_-_118753665
|
2.312
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chrX_+_73641084
|
2.306
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr1_+_203444882
|
2.294
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr15_-_99789585
|
2.201
|
NM_001040655
NM_001040656
NM_001040657
NM_001040658
NM_001040659
NM_001040660
NM_022905
|
TTC23
|
tetratricopeptide repeat domain 23
|
|
chr1_-_154832187
|
2.166
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr10_+_123748688
|
2.064
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr4_-_40859081
|
2.054
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr11_-_119252364
|
1.999
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr4_-_41216455
|
1.963
|
NM_001166050
NM_004307
NM_173075
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr10_+_123923104
|
1.952
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_-_200335988
|
1.825
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr9_+_504661
|
1.665
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr11_-_119234859
|
1.641
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr16_+_77822400
|
1.613
|
NM_020927
|
VAT1L
|
vesicle amine transport protein 1 homolog (T. californica)-like
|
|
chrX_-_103087106
|
1.585
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr3_-_98620452
|
1.583
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr11_-_2162340
|
1.574
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_-_2170792
|
1.564
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_+_65613771
|
1.547
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr5_+_71403040
|
1.508
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr17_+_7184978
|
1.500
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr1_+_65613211
|
1.481
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr12_-_100660752
|
1.480
|
NM_152317
|
DEPDC4
|
DEP domain containing 4
|
|
chr12_-_118406027
|
1.462
|
NM_173598
|
KSR2
|
kinase suppressor of ras 2
|
|
chr9_+_706744
|
1.452
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr11_-_2160199
|
1.449
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr15_+_47476225
|
1.433
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr11_+_94501503
|
1.428
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr12_-_117799410
|
1.421
|
NM_000620
NM_001204218
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chrX_-_112066353
|
1.404
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr1_-_154842725
|
1.395
|
NM_001204087
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr19_+_15218141
|
1.376
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr11_-_66725790
|
1.373
|
NM_000920
NM_001040716
|
PC
|
pyruvate carboxylase
|
|
chr3_-_116164363
|
1.338
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr12_-_49182718
|
1.320
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr2_+_223916861
|
1.301
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr3_-_190040192
|
1.300
|
NM_021101
|
CLDN1
|
claudin 1
|
|
chr15_+_48010685
|
1.276
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr13_+_39261172
|
1.239
|
NM_207361
|
FREM2
|
FRAS1 related extracellular matrix protein 2
|
|
chr6_-_39895317
|
1.231
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr5_+_31799030
|
1.222
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr12_-_49177876
|
1.219
|
NM_015270
|
ADCY6
|
adenylate cyclase 6
|
|
chr18_+_55862577
|
1.204
|
NM_001144970
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr5_-_146889618
|
1.193
|
NM_001197294
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr16_-_70719854
|
1.186
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr14_+_95047730
|
1.174
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr12_-_117742856
|
1.166
|
NM_001204213
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr5_-_146833150
|
1.164
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_-_200329810
|
1.150
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr3_-_57199402
|
1.118
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr2_-_167232464
|
1.114
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr1_+_57110745
|
1.109
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr12_-_117747214
|
1.105
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr15_-_58306185
|
1.105
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr6_-_39902140
|
1.095
|
NM_001075098
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr1_+_163038934
|
1.090
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_-_88974094
|
1.069
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr11_-_66675334
|
1.069
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr13_-_78549663
|
1.066
|
NM_000115
|
EDNRB
|
endothelin receptor type B
|
|
chr20_-_43977023
|
1.033
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr1_+_16091458
|
1.027
|
NM_001024216
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr18_+_55711388
|
1.021
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr12_-_70083055
|
1.018
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr2_-_200322699
|
1.015
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr3_+_39851028
|
1.011
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr1_+_13910251
|
1.011
|
NM_006474
NM_198389
|
PDPN
|
podoplanin
|
|
chr1_-_151804225
|
1.001
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr2_+_5832778
|
0.958
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr15_+_71433787
|
0.958
|
NM_024817
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr16_-_28074776
|
0.951
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chrX_-_31526414
|
0.949
|
NM_004014
|
DMD
|
dystrophin
|
|
chr1_-_43751238
|
0.948
|
NM_182517
NM_001164829
|
C1orf210
|
chromosome 1 open reading frame 210
|
|
chr18_+_55816546
|
0.946
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr11_-_8615412
|
0.941
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr3_+_187871473
|
0.939
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr21_-_27945267
|
0.932
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chrX_-_33229428
|
0.930
|
NM_004006
|
DMD
|
dystrophin
|
|
chr12_-_71031174
|
0.925
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chrX_-_32173585
|
0.925
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr15_-_70994608
|
0.916
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_-_204329043
|
0.906
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr15_-_37393364
|
0.905
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chrX_-_31284946
|
0.897
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chrX_-_32430370
|
0.882
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr3_+_187930720
|
0.870
|
NM_005578
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr2_-_100939194
|
0.862
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr1_-_151798552
|
0.860
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr11_+_33563772
|
0.858
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr3_-_192445344
|
0.858
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr7_-_93204035
|
0.853
|
NM_001164737
NM_001742
|
CALCR
|
calcitonin receptor
|
|
chr4_+_77356252
|
0.851
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr1_+_163038564
|
0.847
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr1_+_203096835
|
0.840
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chrX_-_112084006
|
0.826
|
NM_133265
|
AMOT
|
angiomotin
|
|
chr3_+_153839136
|
0.824
|
NM_001251963
NM_015595
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr9_+_137533643
|
0.816
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr9_+_27109138
|
0.807
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr6_-_57087018
|
0.806
|
NM_016277
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr16_-_66730466
|
0.802
|
NM_178818
NM_181521
|
CMTM4
|
CKLF-like MARVEL transmembrane domain containing 4
|
|
chr1_+_13911966
|
0.794
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr6_-_57086204
|
0.793
|
NM_183227
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr11_+_107461807
|
0.792
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr8_-_81787015
|
0.791
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chrX_-_33146543
|
0.786
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr22_-_24640850
|
0.783
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr13_-_36429798
|
0.778
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr12_-_70093064
|
0.776
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr11_+_56949220
|
0.773
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr10_-_30348433
|
0.764
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr1_+_163041694
|
0.761
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr17_+_64960754
|
0.760
|
NM_014405
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr2_+_166326156
|
0.752
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr12_-_71003622
|
0.749
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr17_+_53342320
|
0.746
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr11_+_8060179
|
0.742
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr3_+_42947649
|
0.742
|
NM_001134656
|
ZNF662
|
zinc finger protein 662
|
|
chr8_+_77593506
|
0.740
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr2_+_27070962
|
0.739
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr18_-_48346297
|
0.736
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr18_+_55888750
|
0.735
|
NM_001144966
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_-_54384436
|
0.735
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chr17_+_8214139
|
0.732
|
NM_025014
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr11_+_12399024
|
0.725
|
NM_018222
|
PARVA
|
parvin, alpha
|
|
chr19_-_11308184
|
0.721
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr3_-_192126837
|
0.717
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr7_-_50861114
|
0.717
|
NM_001001555
|
GRB10
|
growth factor receptor-bound protein 10
|
|
chr10_-_7661613
|
0.709
|
NM_032817
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr1_+_119911395
|
0.709
|
NM_001005783
NM_016527
|
HAO2
|
hydroxyacid oxidase 2 (long chain)
|
|
chr2_-_27718098
|
0.706
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr10_-_100027943
|
0.706
|
NM_032211
|
LOXL4
|
lysyl oxidase-like 4
|
|
chrX_+_68725077
|
0.705
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr12_+_57157091
|
0.704
|
NM_003725
|
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse)
|
|
chr7_-_22396511
|
0.703
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr4_-_46995440
|
0.698
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr17_+_72983723
|
0.696
|
NM_014603
|
CDR2L
|
cerebellar degeneration-related protein 2-like
|
|
chr10_+_71211215
|
0.693
|
NM_012339
|
TSPAN15
|
tetraspanin 15
|
|
chr5_-_11903964
|
0.691
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr16_+_58283819
|
0.688
|
NM_001142302
NM_014157
|
CCDC113
|
coiled-coil domain containing 113
|
|
chr7_-_142583260
|
0.679
|
NM_018646
|
TRPV6
|
transient receptor potential cation channel, subfamily V, member 6
|
|
chr15_-_58358120
|
0.674
|
NM_001206897
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr10_+_128593994
|
0.669
|
NM_001380
|
DOCK1
|
dedicator of cytokinesis 1
|
|
chrX_-_45060081
|
0.667
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr13_+_37248048
|
0.665
|
NM_203451
|
SERTM1
|
serine-rich and transmembrane domain containing 1
|
|
chr5_-_16617085
|
0.657
|
NM_001034850
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr4_+_184020343
|
0.654
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr3_+_42947401
|
0.651
|
NM_207404
|
ZNF662
|
zinc finger protein 662
|
|
chr17_+_47653176
|
0.651
|
NM_007225
|
NXPH3
|
neurexophilin 3
|
|
chr5_-_58652671
|
0.649
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_37392358
|
0.649
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr10_-_128994421
|
0.648
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr10_+_18429605
|
0.643
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr15_-_40633122
|
0.640
|
NM_207380
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr7_+_94539209
|
0.639
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr12_+_26205490
|
0.638
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr2_+_62900985
|
0.633
|
NM_001142615
|
EHBP1
|
EH domain binding protein 1
|
|
chr10_+_18689512
|
0.631
|
NM_201570
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr1_+_202431870
|
0.630
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr11_+_8102691
|
0.630
|
NM_177972
|
TUB
|
tubby homolog (mouse)
|
|
chr12_+_13349601
|
0.627
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_+_26126687
|
0.627
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chrX_+_106161589
|
0.626
|
NM_001171095
|
CLDN2
|
claudin 2
|
|
chr5_-_58571916
|
0.625
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_-_35441099
|
0.624
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr16_-_27899477
|
0.623
|
NM_144675
|
GSG1L
|
GSG1-like
|
|
chr18_-_57364643
|
0.622
|
NM_133459
|
CCBE1
|
collagen and calcium binding EGF domains 1
|
|
chr1_-_55352920
|
0.621
|
NM_014762
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr1_+_78354199
|
0.620
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr13_-_78492906
|
0.619
|
NM_001122659
NM_003991
|
EDNRB
|
endothelin receptor type B
|
|
chrX_+_106163605
|
0.618
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr15_-_56285732
|
0.617
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr1_+_110543548
|
0.614
|
NM_001242674
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr5_+_140723600
|
0.611
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chrX_+_106143393
|
0.606
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr17_+_30593194
|
0.606
|
NM_138328
|
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila)
|
|
chr15_-_63674074
|
0.601
|
NM_001218
NM_206925
|
CA12
|
carbonic anhydrase XII
|
|
chr10_-_44880452
|
0.600
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr8_-_139509064
|
0.600
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr3_+_132757170
|
0.600
|
NM_001136469
NM_023943
|
TMEM108
|
transmembrane protein 108
|
|
chr11_-_35441501
|
0.599
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr18_+_55714457
|
0.598
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_12156584
|
0.596
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr10_-_7708805
|
0.596
|
NM_001001851
NM_030569
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr19_-_42569921
|
0.592
|
NM_002088
|
GRIK5
|
glutamate receptor, ionotropic, kainate 5
|
|
chr4_+_169753105
|
0.591
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr19_-_11305186
|
0.591
|
NM_015493
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr2_-_211035913
|
0.590
|
NM_152519
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chrX_+_37208527
|
0.588
|
NM_000950
NM_001142395
NM_001173486
NM_001173489
NM_001173490
|
PRRG1
|
proline rich Gla (G-carboxyglutamic acid) 1
|
|
chr2_+_62932786
|
0.586
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr15_-_56209305
|
0.582
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr11_+_74862031
|
0.575
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr10_+_18549583
|
0.573
|
NM_000724
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr3_-_87040246
|
0.573
|
NM_016206
|
VGLL3
|
vestigial like 3 (Drosophila)
|
|
chr1_-_117210301
|
0.571
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|