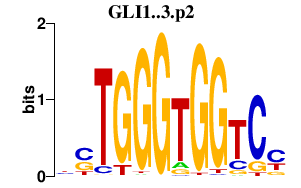
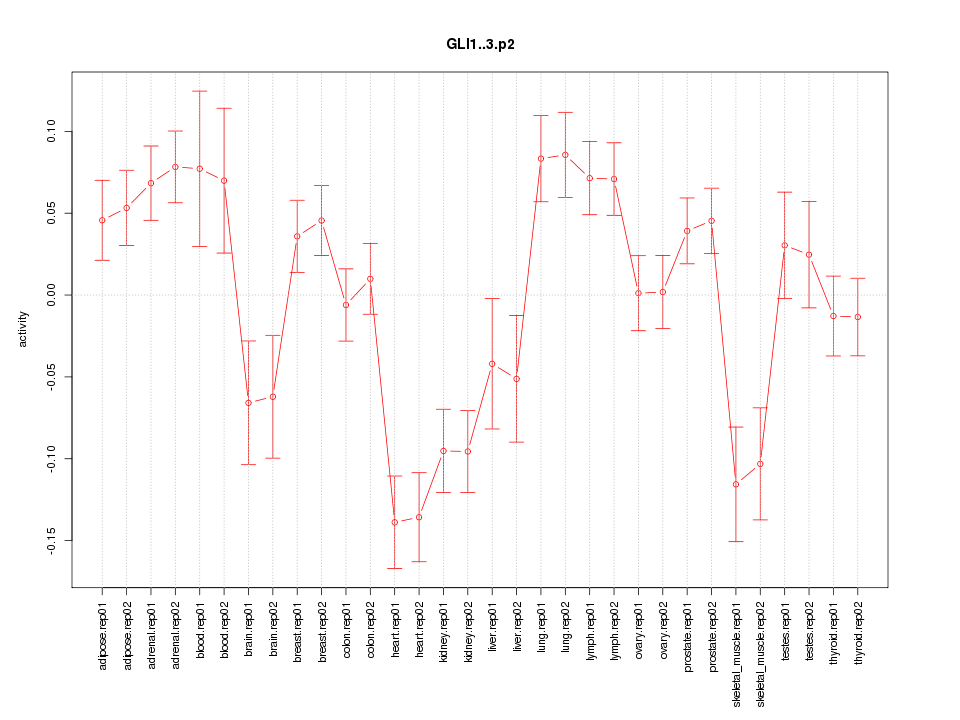
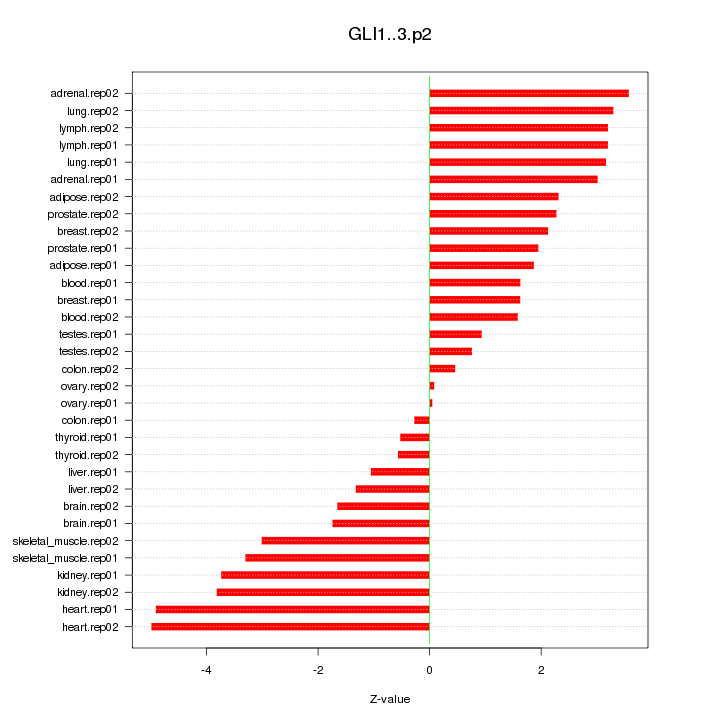
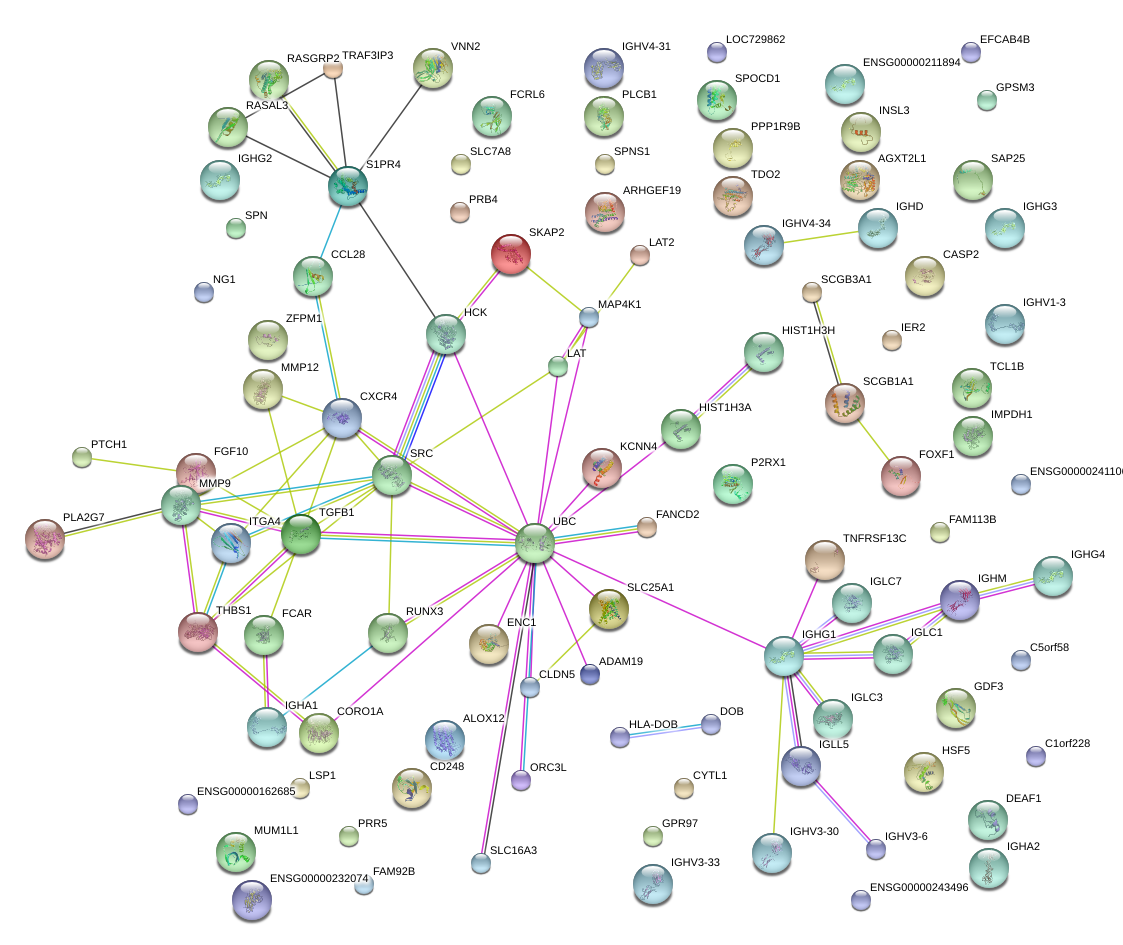
|
chr22_+_22569155
|
6.737
|
|
|
|
|
chr6_-_32784727
|
5.647
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr19_-_15575333
|
5.100
|
NM_022904
|
RASAL3
|
RAS protein activator like 3
|
|
chr14_-_106791054
|
4.855
|
|
IGHV4-31
IGHV3-30
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-30
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr5_-_44388666
|
4.624
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chr5_+_169659945
|
4.573
|
NM_001102609
|
C5orf58
|
chromosome 5 open reading frame 58
|
|
chr22_-_19512001
|
4.198
|
|
CLDN5
|
claudin 5
|
|
chr7_-_100171269
|
3.905
|
NM_001168682
|
SAP25
|
Sin3A-associated protein, 25kDa
|
|
chr15_+_39873276
|
3.826
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr22_-_19511777
|
3.739
|
|
CLDN5
|
claudin 5
|
|
chr22_+_23229959
|
3.575
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr5_-_157002739
|
3.569
|
NM_033274
|
ADAM19
|
ADAM metallopeptidase domain 19
|
|
chr19_-_44285012
|
3.549
|
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr19_+_13263890
|
3.540
|
|
IER2
|
immediate early response 2
|
|
chr17_-_3819692
|
3.520
|
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1
|
|
chr19_-_41859519
|
3.516
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr2_-_136875629
|
3.499
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr14_-_106829736
|
3.413
|
|
IGHA1
IGHD
IGHG3
IGHM
SKAP2
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
src kinase associated phosphoprotein 2
|
|
chr11_-_66084501
|
3.343
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr16_+_29674282
|
3.280
|
NM_001030288
|
SPN
|
sialophorin
|
|
chr19_-_44285407
|
3.271
|
NM_002250
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr14_-_107083398
|
3.235
|
|
IGH@
IGHA1
IGHG1
|
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_45140393
|
3.158
|
NM_001145636
|
C1orf228
|
chromosome 1 open reading frame 228
|
|
chr11_+_62186506
|
3.147
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr16_+_30194925
|
3.116
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr1_+_209941894
|
3.105
|
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr6_-_46703429
|
3.014
|
NM_001168357
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr4_-_5021095
|
3.009
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr9_-_98279246
|
2.942
|
NM_001083602
NM_001083603
|
PTCH1
|
patched 1
|
|
chr16_-_85145995
|
2.929
|
NM_198491
|
FAM92B
|
family with sequence similarity 92, member B
|
|
chr16_+_28996369
|
2.834
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chr20_+_44637539
|
2.806
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr6_-_133084585
|
2.793
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr6_-_32163299
|
2.759
|
NM_022107
|
GPSM3
|
G-protein signaling modulator 3
|
|
chr14_-_106780535
|
2.725
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr19_-_41859767
|
2.719
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr19_-_39108534
|
2.717
|
NM_001042600
NM_007181
|
MAP4K1
|
mitogen-activated protein kinase kinase kinase kinase 1
|
|
chr5_-_180018465
|
2.662
|
NM_052863
|
SCGB3A1
|
secretoglobin, family 3A, member 1
|
|
chr7_+_142985307
|
2.648
|
NM_032982
NM_032983
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr11_+_1892098
|
2.629
|
NM_001013254
NM_001013255
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr17_+_6899383
|
2.573
|
NM_000697
|
ALOX12
|
arachidonate 12-lipoxygenase
|
|
chr6_-_46703042
|
2.542
|
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr17_-_43319100
|
2.482
|
|
|
|
|
chr19_+_3178735
|
2.468
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr19_+_55385548
|
2.460
|
NM_002000
NM_133269
NM_133271
NM_133272
NM_133273
NM_133274
NM_133277
NM_133278
NM_133279
|
FCAR
|
Fc fragment of IgA, receptor for
|
|
chr19_-_41859332
|
2.456
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr16_+_86544104
|
2.454
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr16_+_86544233
|
2.446
|
|
FOXF1
|
forkhead box F1
|
|
chr16_+_30194730
|
2.417
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr6_+_27777713
|
2.394
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr17_-_3819959
|
2.377
|
NM_002558
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1
|
|
chr1_+_159772172
|
2.352
|
NM_001004310
|
FCRL6
|
Fc receptor-like 6
|
|
chr7_+_142985462
|
2.351
|
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase
|
|
chr16_+_57702156
|
2.350
|
NM_170776
|
GPR97
|
G protein-coupled receptor 97
|
|
chr11_-_64511550
|
2.348
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr7_-_128045995
|
2.346
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr7_+_73624252
|
2.319
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr2_+_182321933
|
2.270
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr14_+_96152753
|
2.228
|
NM_004918
|
TCL1B
|
T-cell leukemia/lymphoma 1B
|
|
chr5_-_157002705
|
2.225
|
|
|
|
|
chr3_+_10068139
|
2.222
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr7_+_73624328
|
2.200
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr3_+_10068097
|
2.173
|
NM_001018115
NM_033084
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr12_-_3862353
|
2.147
|
NM_001144958
NM_032680
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr20_+_35974458
|
2.136
|
NM_198291
|
SRC
|
v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian)
|
|
chr12_-_11463353
|
2.128
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr19_-_17932312
|
2.113
|
NM_005543
|
INSL3
|
insulin-like 3 (Leydig cell)
|
|
chr5_-_43412417
|
2.110
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr1_-_32264215
|
2.110
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr1_-_16539103
|
2.106
|
NM_153213
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19
|
|
chr14_-_106714594
|
2.070
|
|
|
|
|
chr11_-_64512265
|
2.060
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr17_-_56565700
|
2.059
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr22_+_45064323
|
2.054
|
NM_001017528
NM_001017529
|
PRR5
|
proline rich 5 (renal)
|
|
chr22_-_42322778
|
2.050
|
NM_052945
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C
|
|
chr17_+_80192774
|
2.048
|
NM_001206951
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr20_+_30639981
|
2.043
|
NM_001172129
NM_001172130
NM_001172131
NM_001172132
NM_001172133
NM_002110
|
HCK
|
hemopoietic cell kinase
|
|
chr12_+_47610244
|
2.035
|
|
FAM113B
|
family with sequence similarity 113, member B
|
|
chr14_-_106321626
|
2.028
|
|
|
|
|
chr12_+_9800510
|
2.021
|
|
LOC374443
|
CLR pseudogene
|
|
chr1_-_25256364
|
2.015
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_-_33793441
|
1.991
|
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr14_-_107259620
|
1.990
|
|
IGHV5-78
|
immunoglobulin heavy variable 5-78 (pseudogene)
|
|
chr12_-_7848324
|
1.989
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr22_-_19166276
|
1.988
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr17_+_7788094
|
1.982
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr17_-_39743085
|
1.975
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr19_-_41858948
|
1.975
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr3_+_38035077
|
1.971
|
NM_015873
|
VILL
|
villin-like
|
|
chr6_-_46703124
|
1.965
|
NM_005084
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma)
|
|
chr7_+_130131921
|
1.964
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_73624347
|
1.948
|
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr3_+_46411632
|
1.947
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr9_-_115652961
|
1.947
|
NM_033051
|
SLC46A2
|
solute carrier family 46, member 2
|
|
chr19_-_33793429
|
1.946
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr2_-_231789940
|
1.906
|
NM_005683
|
GPR55
|
G protein-coupled receptor 55
|
|
chr3_+_10068123
|
1.903
|
|
FANCD2
|
Fanconi anemia, complementation group D2
|
|
chr2_+_182322265
|
1.897
|
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr4_-_109089444
|
1.890
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr12_-_53613883
|
1.867
|
|
RARG
|
retinoic acid receptor, gamma
|
|
chr14_+_77228940
|
1.864
|
|
VASH1
|
vasohibin 1
|
|
chr22_-_19165872
|
1.862
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr3_+_10028579
|
1.858
|
|
LOC442075
|
uncharacterized LOC442075
|
|
chr22_-_19166133
|
1.854
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr3_+_137717657
|
1.852
|
NM_001002026
|
CLDN18
|
claudin 18
|
|
chr5_-_148758787
|
1.848
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr19_+_16435632
|
1.847
|
NM_016270
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr10_+_8097350
|
1.846
|
|
GATA3
|
GATA binding protein 3
|
|
chr14_-_96180402
|
1.842
|
NM_001098725
NM_021966
|
TCL1A
|
T-cell leukemia/lymphoma 1A
|
|
chr12_-_3862217
|
1.837
|
|
EFCAB4B
|
EF-hand calcium binding domain 4B
|
|
chr12_-_45270071
|
1.835
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_+_156123321
|
1.829
|
NM_001193300
NM_022367
NM_001193302
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr19_-_41859830
|
1.825
|
NM_000660
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr7_+_143079354
|
1.824
|
|
ZYX
|
zyxin
|
|
chr22_-_50970542
|
1.811
|
|
ODF3B
|
outer dense fiber of sperm tails 3B
|
|
chr22_+_38035683
|
1.802
|
NM_018957
|
SH3BP1
|
SH3-domain binding protein 1
|
|
chr12_-_108991893
|
1.784
|
NM_181724
|
TMEM119
|
transmembrane protein 119
|
|
chr1_+_248058857
|
1.783
|
NM_001001957
|
OR2W3
|
olfactory receptor, family 2, subfamily W, member 3
|
|
chrX_+_12809473
|
1.782
|
NM_001039091
NM_002765
|
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2
|
|
chr2_+_74648856
|
1.776
|
NM_032118
|
WDR54
|
WD repeat domain 54
|
|
chr15_+_41136622
|
1.775
|
NM_001032367
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1
|
|
chr19_+_38810461
|
1.760
|
NM_004823
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr16_+_27325269
|
1.759
|
|
IL4R
|
interleukin 4 receptor
|
|
chr16_-_3109335
|
1.759
|
|
LOC100507419
|
uncharacterized LOC100507419
|
|
chr1_-_160832466
|
1.753
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr14_+_77228132
|
1.749
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr1_+_17634689
|
1.748
|
NM_012387
|
PADI4
|
peptidyl arginine deiminase, type IV
|
|
chr16_+_50308020
|
1.727
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr1_-_45308603
|
1.723
|
NM_001166292
NM_003738
|
PTCH2
|
patched 2
|
|
chr22_+_39436608
|
1.703
|
NM_001006666
NM_145298
|
APOBEC3F
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F
|
|
chr5_-_168271852
|
1.687
|
|
|
|
|
chr19_+_16435734
|
1.677
|
|
KLF2
|
Kruppel-like factor 2 (lung)
|
|
chr22_-_19166296
|
1.674
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr4_-_164394885
|
1.665
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr19_+_38810540
|
1.662
|
|
KCNK6
|
potassium channel, subfamily K, member 6
|
|
chr1_-_182573393
|
1.639
|
NM_002928
|
RGS16
|
regulator of G-protein signaling 16
|
|
chrX_+_115567746
|
1.636
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr1_+_21766559
|
1.635
|
NM_032264
|
NBPF3
|
neuroblastoma breakpoint family, member 3
|
|
chr1_-_25256767
|
1.632
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr17_-_38721701
|
1.626
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chrY_-_1605999
|
1.616
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chr17_+_72462521
|
1.611
|
NM_007261
|
CD300A
|
CD300a molecule
|
|
chr12_-_53074172
|
1.606
|
NM_006121
|
KRT1
|
keratin 1
|
|
chr17_-_3867585
|
1.598
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr17_+_4802866
|
1.595
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr16_+_67679011
|
1.590
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr14_-_106471439
|
1.581
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr12_-_108991783
|
1.578
|
|
TMEM119
|
transmembrane protein 119
|
|
chr10_-_90712510
|
1.569
|
NM_001613
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr19_+_45418063
|
1.558
|
|
APOC1
|
apolipoprotein C-I
|
|
chr3_-_128840605
|
1.553
|
NM_001204886
NM_001204887
NM_001204888
NM_198490
NM_001204885
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chr12_-_108991832
|
1.543
|
|
TMEM119
|
transmembrane protein 119
|
|
chr7_+_150102376
|
1.537
|
|
|
|
|
chr3_-_128840343
|
1.537
|
|
RAB43
|
RAB43, member RAS oncogene family
|
|
chrX_-_1655999
|
1.537
|
NM_178129
|
P2RY8
CRLF2
|
purinergic receptor P2Y, G-protein coupled, 8
cytokine receptor-like factor 2
|
|
chr19_+_12902286
|
1.529
|
|
|
|
|
chr11_-_128391888
|
1.529
|
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr11_+_46403302
|
1.528
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr5_+_92920592
|
1.521
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_-_78933543
|
1.518
|
NM_000750
|
CHRNB4
|
cholinergic receptor, nicotinic, beta 4
|
|
chr4_-_84030995
|
1.510
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr17_+_80193649
|
1.504
|
NM_001206952
|
SLC16A3
|
solute carrier family 16, member 3 (monocarboxylic acid transporter 4)
|
|
chr7_+_130131172
|
1.502
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr1_+_1115076
|
1.498
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr11_-_61124223
|
1.497
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr16_+_27325244
|
1.493
|
NM_000418
NM_001008699
|
IL4R
|
interleukin 4 receptor
|
|
chr20_+_43343884
|
1.486
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr16_+_28943259
|
1.486
|
NM_001178098
NM_001770
|
CD19
|
CD19 molecule
|
|
chr19_+_860615
|
1.483
|
|
CFD
|
complement factor D (adipsin)
|
|
chr10_+_8096772
|
1.483
|
|
GATA3
|
GATA binding protein 3
|
|
chr1_+_19969722
|
1.479
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr9_-_113800243
|
1.471
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr19_+_54371125
|
1.463
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr16_+_67679058
|
1.462
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr19_-_41859253
|
1.458
|
|
TGFB1
|
transforming growth factor, beta 1
|
|
chr1_-_156675237
|
1.455
|
NM_001878
NM_001199723
|
CRABP2
|
cellular retinoic acid binding protein 2
|
|
chr16_+_28996146
|
1.452
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr17_-_39768939
|
1.451
|
NM_005557
|
KRT16
|
keratin 16
|
|
chrX_+_12809522
|
1.449
|
|
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2
|
|
chr1_+_150122095
|
1.446
|
NM_016274
|
PLEKHO1
|
pleckstrin homology domain containing, family O member 1
|
|
chr16_-_31214054
|
1.446
|
NM_013258
NM_145182
|
PYCARD
|
PYD and CARD domain containing
|
|
chr7_-_92465929
|
1.444
|
NM_001145306
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr20_+_57766074
|
1.443
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr11_+_46403279
|
1.441
|
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr17_-_61920328
|
1.436
|
NM_001098426
|
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr10_+_8096633
|
1.422
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr2_+_219745243
|
1.422
|
NM_025216
|
WNT10A
|
wingless-type MMTV integration site family, member 10A
|
|
chr11_+_46403205
|
1.422
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr10_+_47746919
|
1.418
|
NM_001630
|
ANXA8L2
ANXA8
|
annexin A8-like 2
annexin A8
|
|
chr1_+_161494035
|
1.412
|
NM_002155
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B')
|
|
chr12_-_45269953
|
1.412
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr19_+_36208917
|
1.405
|
NM_014727
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr16_+_31044849
|
1.403
|
|
STX4
|
syntaxin 4
|
|
chr20_+_62328002
|
1.403
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr7_+_73624086
|
1.402
|
NM_014146
NM_032463
NM_032464
|
LAT2
|
linker for activation of T cells family, member 2
|
|
chr7_+_73245192
|
1.402
|
NM_001305
|
CLDN4
|
claudin 4
|
|
chr12_-_45270157
|
1.401
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr19_+_4640028
|
1.400
|
NM_001167942
|
TNFAIP8L1
|
tumor necrosis factor, alpha-induced protein 8-like 1
|
|
chr8_+_75736771
|
1.397
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr19_+_2289996
|
1.383
|
|
|
|