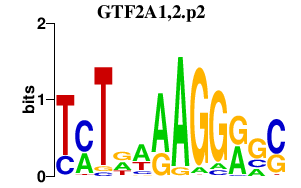
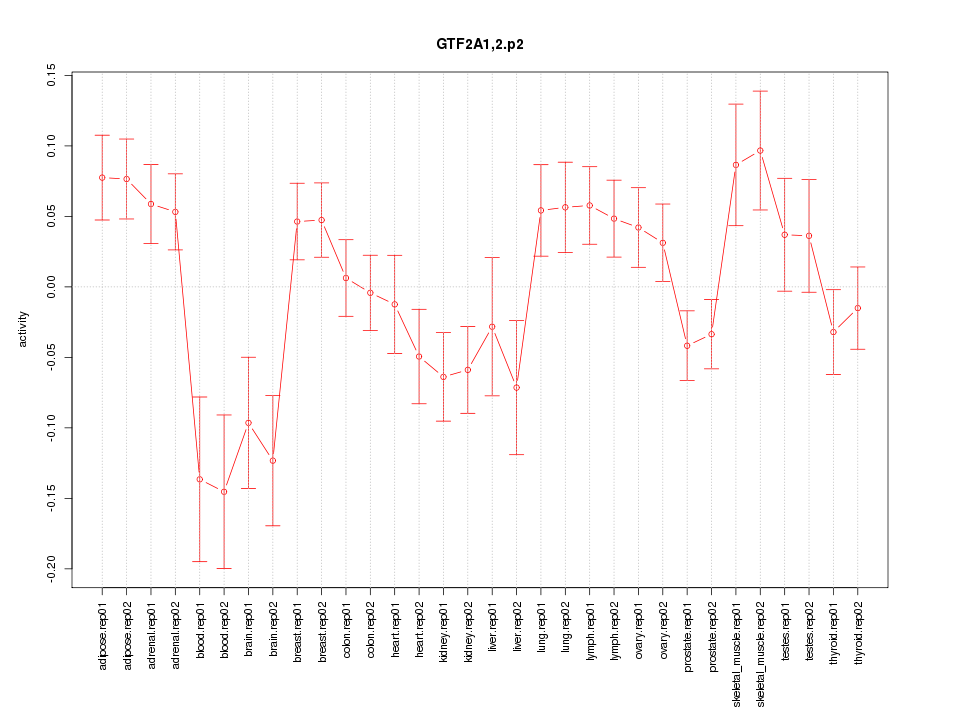
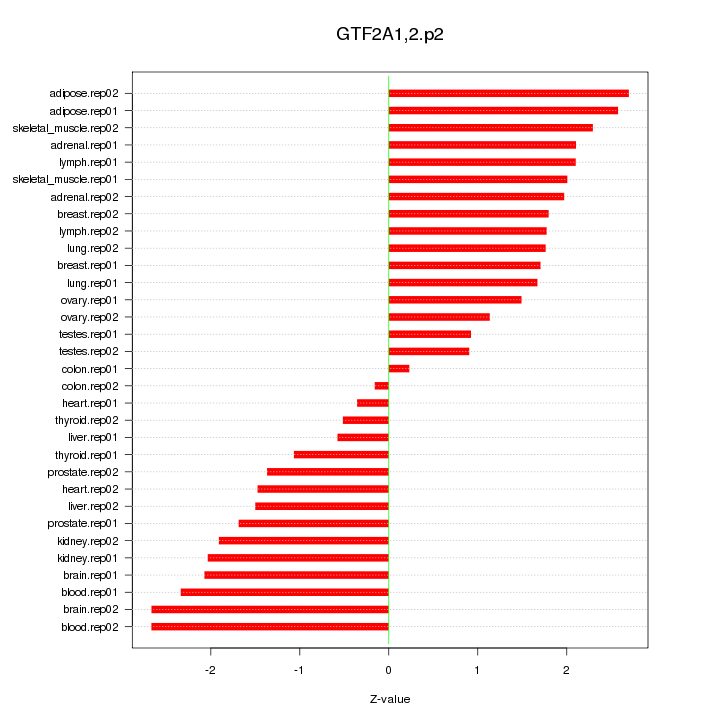
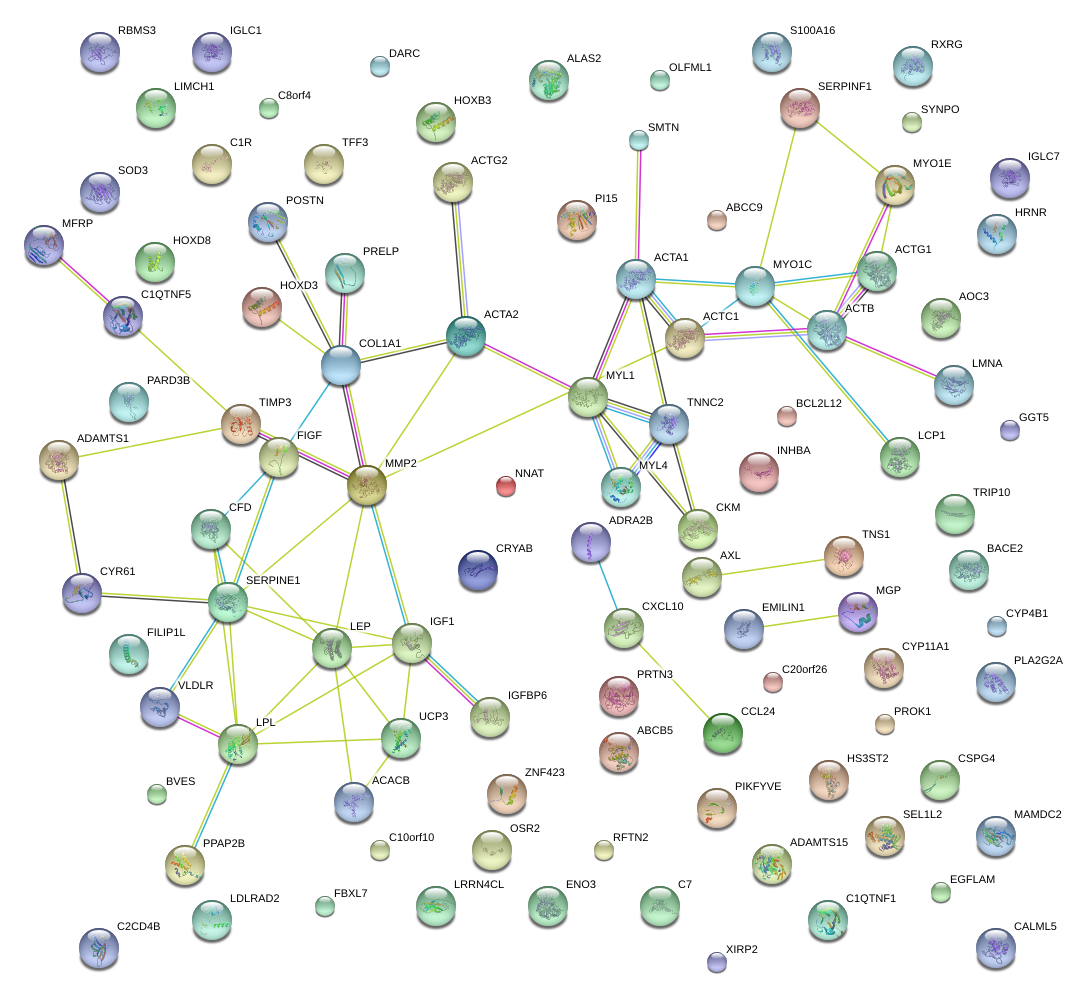
|
chr8_+_99956630
|
5.121
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr12_+_53491417
|
3.863
|
NM_002178
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr2_+_27301434
|
3.842
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr16_+_55515468
|
3.787
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_+_22138757
|
3.769
|
NM_001013693
|
LDLRAD2
|
low density lipoprotein receptor class A domain containing 2
|
|
chr2_+_168043792
|
3.741
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr15_-_76005095
|
3.569
|
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr12_+_53491448
|
3.421
|
|
IGFBP6
|
insulin-like growth factor binding protein 6
|
|
chr15_-_62457481
|
3.404
|
NM_001007595
|
C2CD4B
|
C2 calcium-dependent domain containing 4B
|
|
chr17_-_48278987
|
3.374
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr11_-_62457001
|
3.327
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr12_+_109577201
|
3.306
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr21_-_43735462
|
3.277
|
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr17_-_48278874
|
3.249
|
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr11_+_130318457
|
3.235
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr17_+_41006094
|
3.234
|
|
AOC3
|
amine oxidase, copper containing 3 (vascular adhesion protein 1)
|
|
chr12_-_7245006
|
3.218
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chr15_-_76005179
|
3.125
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chrX_-_15402469
|
3.119
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr17_+_45286713
|
3.087
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr12_-_7245048
|
3.084
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr1_-_20306908
|
3.039
|
NM_000300
NM_001161727
NM_001161728
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr7_+_127881330
|
2.826
|
NM_000230
|
LEP
|
leptin
|
|
chr11_-_62457370
|
2.822
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_153581415
|
2.822
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr2_+_176994421
|
2.816
|
NM_001199747
NM_001199746
NM_019558
|
HOXD8
|
homeobox D8
|
|
chr1_+_159173802
|
2.813
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr2_+_27301693
|
2.736
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr19_+_840970
|
2.725
|
NM_002777
|
PRTN3
|
proteinase 3
|
|
chr10_-_45474210
|
2.679
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr21_-_43735705
|
2.618
|
NM_003226
|
TFF3
|
trefoil factor 3 (intestinal)
|
|
chr20_-_44455932
|
2.614
|
NM_003279
|
TNNC2
|
troponin C type 2 (fast)
|
|
chr12_-_22089627
|
2.611
|
NM_005691
NM_020297
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr2_+_74120132
|
2.539
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr11_-_119217365
|
2.504
|
NM_015645
NM_031433
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr19_+_6739697
|
2.498
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr1_+_86046434
|
2.427
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr17_-_34328607
|
2.301
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chr12_-_15038724
|
2.280
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr8_+_40010986
|
2.258
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr7_+_20686965
|
2.179
|
NM_001163942
NM_001163993
NM_178559
|
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5
|
|
chr1_+_47264669
|
2.166
|
NM_000779
NM_001099772
|
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1
|
|
chr19_+_41725098
|
2.163
|
NM_001699
NM_021913
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr6_-_105584220
|
2.154
|
NM_147147
|
BVES
|
blood vessel epicardial substance
|
|
chr2_+_74120092
|
2.146
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr19_+_41725277
|
2.137
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_-_46651809
|
2.137
|
NM_002146
|
HOXB3
|
homeobox B3
|
|
chr16_-_49860917
|
2.125
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr1_+_203444882
|
2.095
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr2_+_205410465
|
2.077
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr1_-_20306093
|
2.077
|
NM_001161729
|
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid)
|
|
chr11_-_2018568
|
2.063
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_2018446
|
2.006
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_+_7506599
|
1.997
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr2_-_211168315
|
1.995
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr11_-_111782447
|
1.979
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr3_+_29322802
|
1.977
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr15_-_74659926
|
1.970
|
NM_000781
|
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1
|
|
chr11_-_2018334
|
1.957
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_-_165414499
|
1.950
|
NM_006917
|
RXRG
|
retinoid X receptor, gamma
|
|
chr22_-_24640850
|
1.919
|
NM_001099781
NM_001099782
NM_004121
|
GGT5
|
gamma-glutamyltransferase 5
|
|
chr19_+_41725259
|
1.911
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr5_+_38403633
|
1.910
|
NM_182798
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr20_+_20033157
|
1.907
|
NM_015585
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr2_+_177028804
|
1.907
|
NM_006898
|
HOXD3
|
homeobox D3
|
|
chr2_-_198540583
|
1.896
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr1_+_156084579
|
1.896
|
|
LMNA
|
lamin A/C
|
|
chr5_+_40909617
|
1.881
|
|
C7
|
complement component 7
|
|
chrX_-_55057408
|
1.867
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr5_+_15500288
|
1.864
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr20_+_31870940
|
1.856
|
NM_033197
|
BPIFB1
|
BPI fold containing family B, member 1
|
|
chr8_+_75736771
|
1.854
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr16_+_22825806
|
1.853
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr7_-_41742666
|
1.847
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr17_+_4854383
|
1.829
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr4_+_24797084
|
1.813
|
NM_003102
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr5_+_150027837
|
1.799
|
|
SYNPO
|
synaptopodin
|
|
chr11_-_73720281
|
1.778
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr13_-_38172862
|
1.772
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr4_+_24797203
|
1.755
|
|
SOD3
|
superoxide dismutase 3, extracellular
|
|
chr2_-_218808705
|
1.752
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr19_-_45826131
|
1.738
|
|
CKM
|
creatine kinase, muscle
|
|
chr2_-_96781887
|
1.731
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr19_+_41725131
|
1.726
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr19_+_41725135
|
1.723
|
|
AXL
|
AXL receptor tyrosine kinase
|
|
chr17_+_1665233
|
1.721
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr20_+_36149606
|
1.703
|
NM_005386
NM_181689
|
NNAT
|
neuronatin
|
|
chr17_+_77030471
|
1.674
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr1_-_57045214
|
1.661
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr17_+_1665342
|
1.660
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr21_-_28217692
|
1.659
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr17_-_1389013
|
1.652
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr19_-_45826232
|
1.651
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr9_+_72658496
|
1.637
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr7_-_75443026
|
1.621
|
NM_002991
|
CCL24
|
chemokine (C-C motif) ligand 24
|
|
chr1_-_57044980
|
1.616
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr22_+_22758538
|
1.614
|
|
|
|
|
chr1_-_57045129
|
1.606
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr12_-_102874373
|
1.604
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr4_+_41614911
|
1.604
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_110993748
|
1.603
|
NM_032414
|
PROK1
|
prokineticin 1
|
|
chr22_+_31487315
|
1.594
|
|
SMTN
|
smoothelin
|
|
chr1_-_57045235
|
1.573
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr1_+_156084565
|
1.566
|
|
LMNA
|
lamin A/C
|
|
chr12_-_102872329
|
1.563
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr21_+_42540092
|
1.562
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr10_-_5541401
|
1.557
|
NM_017422
|
CALML5
|
calmodulin-like 5
|
|
chr19_+_859673
|
1.553
|
|
CFD
|
complement factor D (adipsin)
|
|
chr20_-_13971256
|
1.550
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr11_-_2019070
|
1.545
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr21_+_42540054
|
1.541
|
|
BACE2
|
beta-site APP-cleaving enzyme 2
|
|
chr19_+_859658
|
1.540
|
NM_001928
|
CFD
|
complement factor D (adipsin)
|
|
chr9_+_2621740
|
1.537
|
NM_001018056
NM_003383
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr1_+_156084541
|
1.536
|
|
LMNA
|
lamin A/C
|
|
chr22_+_33196801
|
1.535
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr5_+_150020201
|
1.521
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr3_-_99594915
|
1.509
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr8_+_19796581
|
1.508
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr7_+_100770378
|
1.480
|
NM_000602
NM_001165413
|
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1
|
|
chr5_+_140566655
|
1.479
|
NM_019119
|
PCDHB9
|
protocadherin beta 9
|
|
chr9_+_2622134
|
1.478
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr19_+_840963
|
1.475
|
|
PRTN3
|
proteinase 3
|
|
chr6_-_105584542
|
1.470
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr17_-_66951381
|
1.464
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr17_+_48133357
|
1.464
|
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr11_-_5531152
|
1.460
|
NM_017481
|
UBQLN3
|
ubiquilin 3
|
|
chr1_-_168105620
|
1.458
|
NM_153832
|
GPR161
|
G protein-coupled receptor 161
|
|
chr2_+_95537177
|
1.457
|
NM_144705
|
TEKT4
|
tektin 4
|
|
chr6_-_85472312
|
1.452
|
|
TBX18
|
T-box 18
|
|
chr2_-_110873533
|
1.427
|
|
MALL
|
mal, T-cell differentiation protein-like
|
|
chr13_-_110959441
|
1.424
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr5_+_40909487
|
1.424
|
|
C7
|
complement component 7
|
|
chr16_+_215972
|
1.423
|
NM_001003938
|
HBM
|
hemoglobin, mu
|
|
chr11_-_64825992
|
1.423
|
NM_005468
|
NAALADL1
|
N-acetylated alpha-linked acidic dipeptidase-like 1
|
|
chr3_-_149375584
|
1.416
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr9_+_124043719
|
1.414
|
|
GSN
|
gelsolin
|
|
chr3_-_78666943
|
1.414
|
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr5_+_40909558
|
1.414
|
NM_000587
|
C7
|
complement component 7
|
|
chr22_+_22676814
|
1.411
|
|
|
|
|
chr9_+_72658735
|
1.399
|
|
MAMDC2
|
MAM domain containing 2
|
|
chrX_+_99899162
|
1.397
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr15_+_74466024
|
1.389
|
NM_005545
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr12_+_54378929
|
1.389
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr16_+_811049
|
1.383
|
NM_013404
|
MSLN
|
mesothelin
|
|
chr21_+_19617145
|
1.381
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr8_+_19796730
|
1.379
|
|
LPL
|
lipoprotein lipase
|
|
chr1_+_53527822
|
1.378
|
NM_001199081
|
PODN
|
podocan
|
|
chr1_+_156084458
|
1.377
|
NM_005572
NM_170707
NM_170708
|
LMNA
|
lamin A/C
|
|
chr1_+_159141376
|
1.372
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr4_+_41614933
|
1.371
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr20_-_3662635
|
1.357
|
NM_025220
NM_153202
|
ADAM33
|
ADAM metallopeptidase domain 33
|
|
chr12_-_8088629
|
1.355
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr19_-_55660557
|
1.352
|
NM_001126132
NM_001126133
NM_003283
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr13_+_111154013
|
1.345
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr2_+_205410556
|
1.340
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chrX_+_99899388
|
1.332
|
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr12_-_56106061
|
1.331
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr17_+_61554420
|
1.311
|
NM_000789
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr19_-_42947050
|
1.302
|
NM_198477
|
CXCL17
|
chemokine (C-X-C motif) ligand 17
|
|
chr17_+_48133331
|
1.279
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr6_-_123957941
|
1.274
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr7_-_99698916
|
1.272
|
|
MCM7
|
minichromosome maintenance complex component 7
|
|
chr9_+_136243226
|
1.267
|
NM_153710
|
C9orf96
|
chromosome 9 open reading frame 96
|
|
chr8_+_144295067
|
1.265
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr16_+_31539202
|
1.264
|
NM_016633
|
AHSP
|
alpha hemoglobin stabilizing protein
|
|
chr13_-_40177200
|
1.263
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr16_+_22825481
|
1.262
|
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr7_+_116395440
|
1.257
|
|
|
|
|
chr5_-_134369571
|
1.256
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr1_+_53527723
|
1.255
|
NM_001199080
NM_001199082
NM_153703
|
PODN
|
podocan
|
|
chr12_+_13349601
|
1.250
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_-_27339326
|
1.248
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr10_+_88426538
|
1.246
|
|
LDB3
|
LIM domain binding 3
|
|
chr12_+_130647002
|
1.246
|
NM_007197
|
FZD10
|
frizzled family receptor 10
|
|
chr2_+_56411123
|
1.245
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr3_-_48130283
|
1.239
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr17_+_37821598
|
1.239
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr11_-_33744272
|
1.237
|
NM_001127223
NM_001127225
NM_001127226
|
CD59
|
CD59 molecule, complement regulatory protein
|
|
chr7_+_6793739
|
1.228
|
NM_173565
NM_001099697
|
RSPH10B
RSPH10B2
|
radial spoke head 10 homolog B (Chlamydomonas)
radial spoke head 10 homolog B2 (Chlamydomonas)
|
|
chr18_-_22931970
|
1.214
|
|
ZNF521
|
zinc finger protein 521
|
|
chr6_-_112575880
|
1.204
|
|
LAMA4
|
laminin, alpha 4
|
|
chr1_+_203444955
|
1.199
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr6_-_29527477
|
1.199
|
|
UBD
|
ubiquitin D
|
|
chr8_+_37553269
|
1.194
|
NM_025069
|
ZNF703
|
zinc finger protein 703
|
|
chr3_+_42700837
|
1.192
|
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr9_+_2622099
|
1.190
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr4_-_76957139
|
1.189
|
NM_005409
|
CXCL11
|
chemokine (C-X-C motif) ligand 11
|
|
chr18_-_53303129
|
1.175
|
NM_001243226
|
TCF4
|
transcription factor 4
|
|
chr21_+_47546427
|
1.171
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr14_-_107122358
|
1.167
|
|
|
|
|
chr1_-_169703178
|
1.166
|
NM_000450
|
SELE
|
selectin E
|
|
chr22_+_23029175
|
1.162
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr18_-_22932109
|
1.161
|
NM_015461
|
ZNF521
|
zinc finger protein 521
|
|
chrX_-_139866582
|
1.159
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chrX_-_15333726
|
1.141
|
NM_001201583
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr1_+_171060017
|
1.137
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr14_+_105953531
|
1.137
|
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr1_+_84768333
|
1.134
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr21_-_28217272
|
1.131
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr3_+_140396865
|
1.131
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|