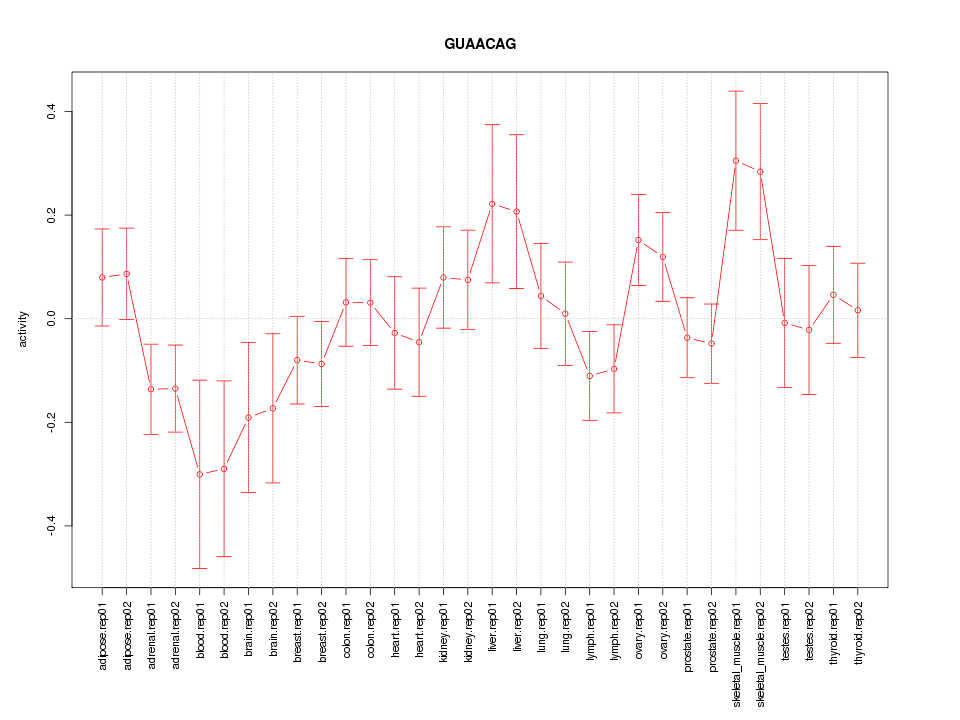
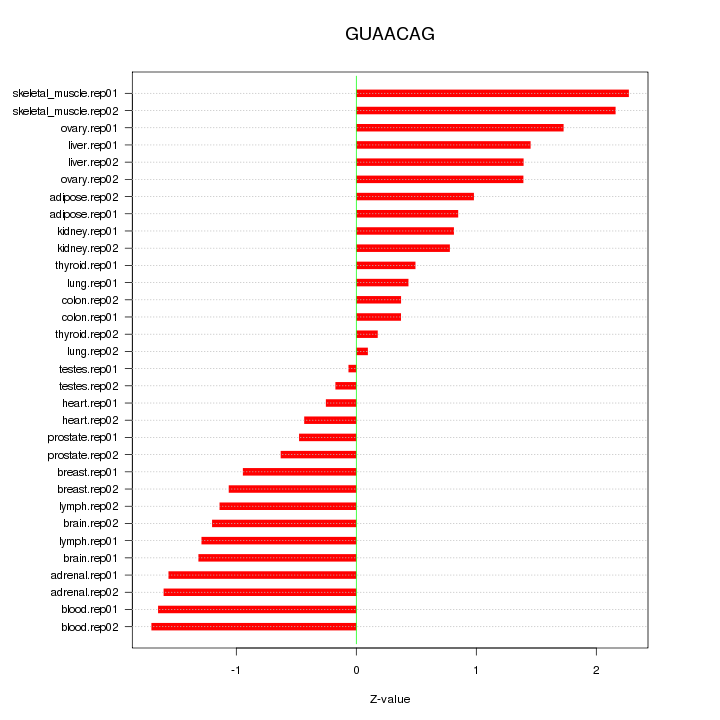
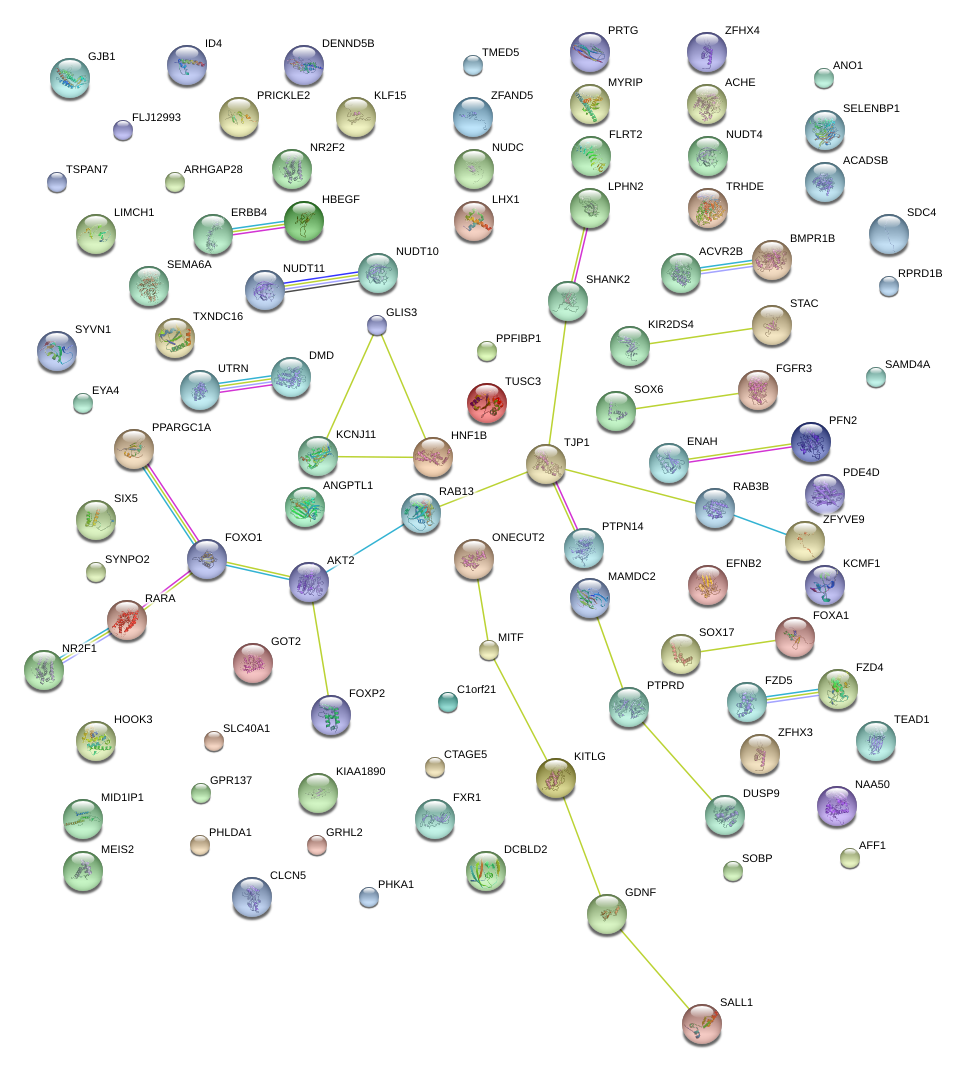
|
chr17_-_36105005
|
3.223
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr6_+_133562488
|
3.137
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr11_+_12695851
|
3.094
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_-_126076057
|
2.870
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr8_+_55370494
|
2.641
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr15_+_96875793
|
2.512
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_+_96873845
|
2.496
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr15_+_96869156
|
2.326
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_178840079
|
2.260
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr11_-_17410205
|
2.235
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr11_-_17410863
|
2.217
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr1_+_82266081
|
2.216
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr14_-_38064440
|
2.200
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr11_-_16424391
|
2.087
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr15_+_96876568
|
1.999
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr11_-_16497917
|
1.924
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr19_-_46272496
|
1.896
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr16_-_73092533
|
1.859
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr11_-_16430425
|
1.828
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_-_225840660
|
1.819
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chrX_+_70443055
|
1.813
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chrX_-_71933709
|
1.807
|
NM_001122670
NM_001172436
NM_002637
|
PHKA1
|
phosphorylase kinase, alpha 1 (muscle)
|
|
chr11_+_69924407
|
1.772
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr20_-_43977023
|
1.765
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr4_-_23891608
|
1.740
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr14_+_85996454
|
1.717
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chrX_+_49687212
|
1.690
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr13_-_107187312
|
1.652
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr2_-_208634051
|
1.611
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chrX_+_38420690
|
1.607
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr1_-_214724974
|
1.529
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr9_+_72658496
|
1.488
|
NM_153267
|
MAMDC2
|
MAM domain containing 2
|
|
chr1_-_156051788
|
1.473
|
NM_001093725
|
MEX3A
|
mex-3 homolog A (C. elegans)
|
|
chr3_+_38495778
|
1.446
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr12_+_27677044
|
1.431
|
NM_001198915
NM_001198916
NM_003622
NM_177444
|
PPFIBP1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr2_+_85198176
|
1.411
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr9_-_4300028
|
1.386
|
NM_001042413
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr2_-_190445521
|
1.385
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr3_-_149688638
|
1.381
|
NM_002628
NM_053024
|
PFN2
|
profilin 2
|
|
chr8_+_77593506
|
1.370
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr3_+_69915374
|
1.353
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_102504661
|
1.332
|
NM_024915
|
GRHL2
|
grainyhead-like 2 (Drosophila)
|
|
chr3_+_69788562
|
1.327
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr9_-_8733893
|
1.311
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr15_-_56035176
|
1.280
|
NM_173814
|
PRTG
|
protogenin
|
|
chr6_+_19837599
|
1.255
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr16_-_73082169
|
1.250
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr16_-_58768132
|
1.230
|
NM_002080
|
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2)
|
|
chr1_-_93646245
|
1.216
|
NM_001167830
NM_016040
|
TMED5
|
transmembrane emp24 protein transport domain containing 5
|
|
chr12_-_76425375
|
1.214
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chrX_+_49832214
|
1.177
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr2_-_213403280
|
1.155
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr5_-_139726173
|
1.139
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr16_-_51185150
|
1.130
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chrX_+_38660653
|
1.097
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr7_-_100493481
|
1.092
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr8_+_42752019
|
1.056
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr16_-_51184507
|
1.051
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr18_+_55102777
|
1.042
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr4_+_95679075
|
1.038
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr3_+_69812620
|
1.026
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr3_+_39851028
|
1.024
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr14_+_55034329
|
1.015
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr5_-_58571916
|
1.014
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_-_113465101
|
1.011
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr1_-_52456292
|
1.009
|
NM_002867
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr4_+_119809978
|
0.992
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr9_-_4152182
|
0.980
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr9_-_10612722
|
0.966
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr1_-_153958688
|
0.958
|
NM_002870
|
RAB13
|
RAB13, member RAS oncogene family
|
|
chr14_+_39734475
|
0.952
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr3_+_69812961
|
0.950
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_-_115910406
|
0.944
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr11_-_64901995
|
0.942
|
NM_032431
NM_172230
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin
|
|
chr1_+_184356128
|
0.938
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr8_+_15397595
|
0.931
|
NM_006765
NM_178234
|
TUSC3
|
tumor suppressor candidate 3
|
|
chr12_+_72666462
|
0.925
|
NM_013381
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme
|
|
chr4_+_87856152
|
0.923
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr5_-_59064437
|
0.922
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr19_-_40791291
|
0.920
|
NM_001243027
NM_001243028
NM_001626
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr18_+_6834431
|
0.896
|
NM_001010000
|
ARHGAP28
|
Rho GTPase activating protein 28
|
|
chr4_+_41614911
|
0.891
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr13_-_41240607
|
0.889
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr4_+_41362750
|
0.887
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_37835928
|
0.877
|
NM_001190468
NM_001190469
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr11_-_70935807
|
0.855
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chrX_-_31526414
|
0.851
|
NM_004014
|
DMD
|
dystrophin
|
|
chr15_-_37393364
|
0.825
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr3_-_98620452
|
0.824
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr5_-_58652671
|
0.817
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_-_70507911
|
0.816
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr5_-_59189620
|
0.798
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_30114536
|
0.791
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chrX_-_32173585
|
0.787
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chrX_-_32430370
|
0.783
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr7_+_114055048
|
0.775
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr4_-_83483099
|
0.775
|
NM_001080506
|
TMEM150C
|
transmembrane protein 150C
|
|
chr1_+_27248212
|
0.770
|
NM_006600
|
NUDC
|
nuclear distribution gene C homolog (A. nidulans)
|
|
chr3_+_36421944
|
0.759
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chrX_-_31284946
|
0.752
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr12_-_88974094
|
0.751
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr10_+_124768401
|
0.748
|
NM_001609
|
ACADSB
|
acyl-CoA dehydrogenase, short/branched chain
|
|
chr5_-_37835592
|
0.737
|
NM_199231
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr11_-_86666211
|
0.732
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr5_-_37839781
|
0.728
|
NM_000514
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chrX_+_70435048
|
0.725
|
NM_001097642
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr1_+_52608045
|
0.719
|
NM_004799
NM_007323
NM_007324
|
ZFYVE9
|
zinc finger, FYVE domain containing 9
|
|
chr20_+_36661849
|
0.713
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr3_-_64211111
|
0.707
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chrX_-_33357725
|
0.699
|
NM_000109
|
DMD
|
dystrophin
|
|
chr4_+_1795004
|
0.697
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr3_+_180630054
|
0.690
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr12_+_93771607
|
0.684
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr5_-_58334936
|
0.677
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chrX_+_152907892
|
0.672
|
NM_001395
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr12_-_31743822
|
0.670
|
NM_144973
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr17_+_38465422
|
0.670
|
NM_000964
|
RARA
|
retinoic acid receptor, alpha
|
|
chr6_+_107811272
|
0.667
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr9_-_74979829
|
0.660
|
NM_001102420
NM_001102421
NM_006007
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chrX_-_33229428
|
0.657
|
NM_004006
|
DMD
|
dystrophin
|
|
chr17_+_38498270
|
0.656
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr17_+_35294771
|
0.653
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr21_-_16437125
|
0.645
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr16_-_12009804
|
0.638
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr7_-_94285477
|
0.623
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr5_-_141704575
|
0.622
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr4_+_87928083
|
0.608
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr12_-_23737507
|
0.600
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr17_-_8534008
|
0.598
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr3_-_122233781
|
0.594
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr12_-_102872329
|
0.577
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_+_177019196
|
0.575
|
NM_017510
|
TMED9
|
transmembrane emp24 protein transport domain containing 9
|
|
chr3_+_69985750
|
0.562
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chrX_-_33146543
|
0.560
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr2_-_174828775
|
0.550
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr5_-_41510627
|
0.548
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr6_+_43445255
|
0.523
|
NM_001146016
NM_001146018
NM_001146019
NM_001146020
NM_080604
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr11_+_22689653
|
0.521
|
NM_177553
|
GAS2
|
growth arrest-specific 2
|
|
chr10_-_13390189
|
0.520
|
NM_001195602
NM_001195604
NM_012247
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr9_+_35161950
|
0.516
|
NM_006377
|
UNC13B
|
unc-13 homolog B (C. elegans)
|
|
chr2_-_158732341
|
0.507
|
NM_001111067
|
ACVR1
|
activin A receptor, type I
|
|
chr3_+_148709140
|
0.502
|
NM_001184720
NM_001184721
NM_004130
|
GYG1
|
glycogenin 1
|
|
chr14_-_100070442
|
0.501
|
NM_001144995
|
CCDC85C
|
coiled-coil domain containing 85C
|
|
chr12_-_24715349
|
0.499
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr15_-_37392358
|
0.492
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_158731622
|
0.486
|
NM_001105
|
ACVR1
|
activin A receptor, type I
|
|
chr12_+_65218351
|
0.484
|
NM_015279
|
TBC1D30
|
TBC1 domain family, member 30
|
|
chr10_-_33623771
|
0.482
|
NM_001024628
NM_001024629
NM_001244972
NM_001244973
NM_003873
|
NRP1
|
neuropilin 1
|
|
chr5_-_59783885
|
0.480
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr4_-_74904329
|
0.479
|
NM_002090
|
CXCL3
|
chemokine (C-X-C motif) ligand 3
|
|
chr7_-_139876718
|
0.474
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr10_+_31608097
|
0.470
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chrX_-_135338640
|
0.466
|
NM_001173516
|
MAP7D3
|
MAP7 domain containing 3
|
|
chr8_-_33370552
|
0.458
|
NM_025115
NM_001102401
|
TTI2
|
TELO2 interacting protein 2
|
|
chr13_-_45992474
|
0.455
|
NM_001010875
|
SLC25A30
|
solute carrier family 25, member 30
|
|
chr4_+_184020343
|
0.449
|
NM_024949
|
WWC2
|
WW and C2 domain containing 2
|
|
chr3_+_147127151
|
0.440
|
NM_003412
|
ZIC1
|
Zic family member 1
|
|
chr12_+_107349539
|
0.435
|
NM_152261
|
C12orf23
|
chromosome 12 open reading frame 23
|
|
chr1_+_112938799
|
0.432
|
NM_018704
|
CTTNBP2NL
|
CTTNBP2 N-terminal like
|
|
chr6_-_83903632
|
0.413
|
NM_001199917
|
PGM3
|
phosphoglucomutase 3
|
|
chr8_-_38326351
|
0.413
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_-_76934508
|
0.412
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr3_+_197476423
|
0.410
|
NM_001011537
|
FYTTD1
|
forty-two-three domain containing 1
|
|
chr16_+_47495166
|
0.408
|
NM_000293
NM_001031835
|
PHKB
|
phosphorylase kinase, beta
|
|
chr16_-_12010518
|
0.396
|
NM_001130007
|
GSPT1
|
G1 to S phase transition 1
|
|
chr17_-_33390676
|
0.394
|
NM_001017368
|
RFFL
|
ring finger and FYVE-like domain containing 1
|
|
chr11_+_22696318
|
0.393
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chr12_-_54020081
|
0.392
|
NM_001130060
NM_001206682
NM_006856
|
ATF7
|
activating transcription factor 7
|
|
chr22_+_41253055
|
0.387
|
NM_001204827
NM_022098
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr11_+_107461807
|
0.385
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr3_+_197476623
|
0.383
|
NM_032288
|
FYTTD1
|
forty-two-three domain containing 1
|
|
chr11_-_115375065
|
0.381
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr17_-_1359410
|
0.381
|
NM_005206
NM_016823
|
CRK
|
v-crk sarcoma virus CT10 oncogene homolog (avian)
|
|
chr17_+_38474472
|
0.380
|
NM_001145301
NM_001145302
|
RARA
|
retinoic acid receptor, alpha
|
|
chr10_+_114709963
|
0.379
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_+_32280154
|
0.372
|
NM_178868
|
CMTM8
|
CKLF-like MARVEL transmembrane domain containing 8
|
|
chr20_-_25604583
|
0.366
|
NM_152667
|
NANP
|
N-acetylneuraminic acid phosphatase
|
|
chr19_+_41903693
|
0.365
|
NM_000709
NM_001164783
|
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide
|
|
chr8_-_97172943
|
0.364
|
NM_001001557
|
GDF6
|
growth differentiation factor 6
|
|
chr8_-_38325241
|
0.363
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_-_88119604
|
0.361
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_58906968
|
0.361
|
NM_147189
|
FAM110B
|
family with sequence similarity 110, member B
|
|
chr17_+_66508098
|
0.349
|
NM_212471
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1)
|
|
chr14_+_77564570
|
0.342
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chr20_-_524335
|
0.342
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr3_-_196242190
|
0.341
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr12_+_123868608
|
0.334
|
NM_020382
|
SETD8
|
SET domain containing (lysine methyltransferase) 8
|
|
chr20_-_60640842
|
0.327
|
NM_003185
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
|
|
chr7_+_138916230
|
0.325
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr16_+_69599868
|
0.322
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr10_+_31610063
|
0.321
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chrX_+_119737743
|
0.320
|
NM_014060
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chrX_+_38663072
|
0.318
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr7_-_152133071
|
0.317
|
NM_170606
|
MLL3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr3_-_118864881
|
0.313
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr11_+_22688156
|
0.312
|
NM_001143830
|
GAS2
|
growth arrest-specific 2
|
|
chr12_-_24102563
|
0.311
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_-_122406994
|
0.309
|
NM_001142273
NM_001142274
NM_001207051
NM_015282
|
CLASP1
|
cytoplasmic linker associated protein 1
|
|
chr11_-_59436488
|
0.304
|
NM_152716
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr11_+_111473099
|
0.297
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|