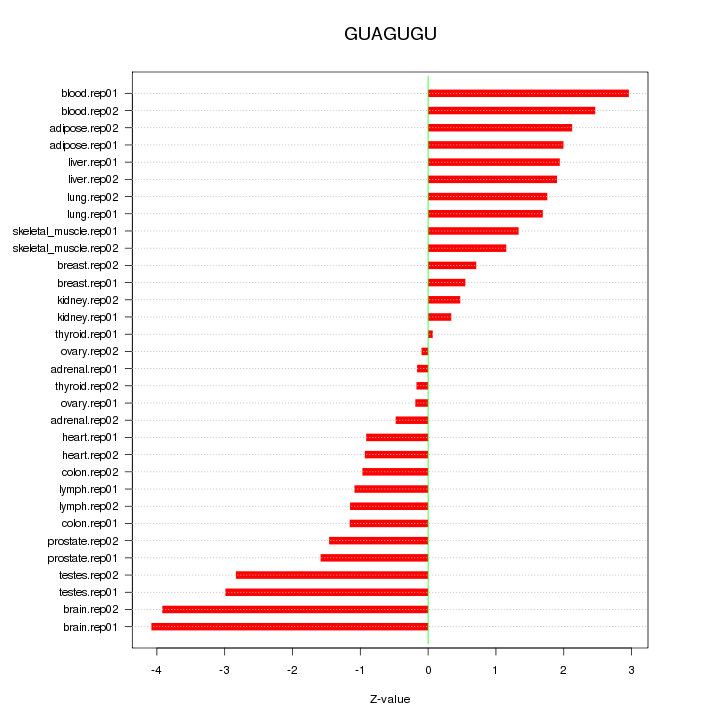
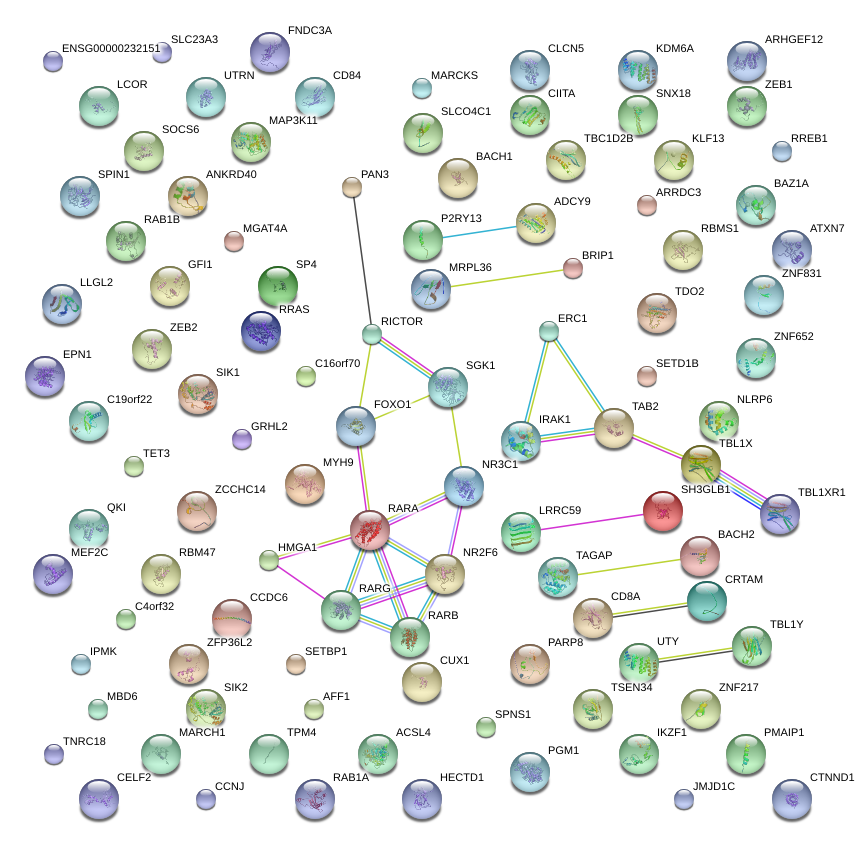
|
chr5_-_101631965
|
7.125
|
NM_180991
|
SLCO4C1
|
solute carrier organic anion transporter family, member 4C1
|
|
chr7_+_50344255
|
6.474
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chrX_+_49832214
|
5.509
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr4_-_40631846
|
5.334
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr6_-_91006580
|
5.275
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr6_-_91006460
|
5.246
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr21_-_44846927
|
4.679
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr5_+_49962771
|
4.672
|
NM_001178056
NM_024615
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr20_+_57766074
|
4.606
|
NM_178457
|
ZNF831
|
zinc finger protein 831
|
|
chr21_+_30671736
|
4.595
|
NM_206866
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr2_-_145277879
|
4.436
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr14_-_35344852
|
4.339
|
NM_013448
NM_182648
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A
|
|
chrX_+_9432980
|
4.334
|
NM_005647
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_-_52199635
|
4.289
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr1_-_92951593
|
4.236
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr1_-_92952429
|
4.164
|
NM_005263
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr12_+_57916607
|
4.096
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr5_-_39074494
|
3.908
|
NM_152756
|
RICTOR
|
RPTOR independent companion of MTOR, complex 2
|
|
chr1_-_29450406
|
3.768
|
NM_001003682
|
TMEM200B
|
transmembrane protein 200B
|
|
chrX_+_49687212
|
3.756
|
NM_001127898
NM_001127899
|
CLCN5
|
chloride channel 5
|
|
chr2_-_161350304
|
3.643
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr5_-_90679115
|
3.620
|
NM_020801
|
ARRDC3
|
arrestin domain containing 3
|
|
chr10_-_60027630
|
3.601
|
NM_152230
|
IPMK
|
inositol polyphosphate multikinase
|
|
chr11_+_111473099
|
3.575
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chrX_-_108976509
|
3.559
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr13_-_41240607
|
3.551
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr6_+_144612872
|
3.470
|
NM_007124
|
UTRN
|
utrophin
|
|
chr10_-_65028825
|
3.461
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr1_-_92949355
|
3.455
|
NM_001127215
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr12_+_122242580
|
3.447
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|
|
chr7_+_101460881
|
3.445
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr1_-_29449012
|
3.435
|
NM_001171868
|
TMEM200B
|
transmembrane protein 200B
|
|
chr15_-_78369825
|
3.403
|
NM_015079
NM_144572
|
TBC1D2B
|
TBC1 domain family, member 2B
|
|
chr4_-_40517913
|
3.386
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr15_+_31619043
|
3.252
|
NM_015995
|
KLF13
|
Kruppel-like factor 13
|
|
chr2_-_87035518
|
3.232
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr3_+_63884074
|
3.231
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr11_+_122709205
|
3.222
|
NM_019604
|
CRTAM
|
cytotoxic and regulatory T cell molecule
|
|
chr5_+_49961732
|
3.176
|
NM_001178055
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr2_-_87018802
|
3.142
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr2_-_99347588
|
3.139
|
NM_012214
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr4_+_87856152
|
3.050
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr6_+_7107986
|
3.034
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr7_-_5463176
|
3.005
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr11_+_57529233
|
2.978
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chrX_-_153285334
|
2.956
|
NM_001025242
NM_001025243
NM_001569
|
IRAK1
|
interleukin-1 receptor-associated kinase 1
|
|
chr21_+_30671217
|
2.916
|
NM_001186
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr6_+_163835668
|
2.887
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr16_-_4166185
|
2.840
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr18_+_42260774
|
2.809
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr6_+_34206567
|
2.776
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_-_142783975
|
2.732
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_-_160549271
|
2.708
|
NM_001184879
NM_001184881
NM_001184882
NM_003874
|
CD84
|
CD84 molecule
|
|
chr17_-_47439326
|
2.695
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr4_+_87928083
|
2.688
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr18_+_57567191
|
2.625
|
NM_021127
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr4_-_164534656
|
2.586
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr10_+_98592658
|
2.505
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr16_+_10971038
|
2.489
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr6_+_114178516
|
2.464
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr16_+_28986024
|
2.462
|
NM_001142448
NM_001142449
NM_001142450
NM_001142451
NM_032038
|
SPIN1
SPNS1
|
spindlin 1
spinster homolog 1 (Drosophila)
|
|
chr3_+_63953419
|
2.457
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr5_-_142815076
|
2.410
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr10_+_97803064
|
2.395
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr3_-_151047309
|
2.332
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr6_+_7107799
|
2.327
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr13_+_49684388
|
2.304
|
NM_014923
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr6_-_159466019
|
2.252
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr2_+_74273449
|
2.252
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr12_+_1100352
|
2.239
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr2_-_43453737
|
2.235
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr19_+_16187047
|
2.219
|
NM_003290
|
TPM4
|
tropomyosin 4
|
|
chr22_-_36784055
|
2.174
|
NM_002473
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr18_+_67956136
|
2.162
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr19_-_17356022
|
2.156
|
NM_005234
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6
|
|
chr12_-_53625834
|
2.108
|
NM_000966
NM_001243730
NM_001243731
|
RARG
|
retinoic acid receptor, gamma
|
|
chr11_-_65381641
|
2.100
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr5_+_53813494
|
2.072
|
NM_001145427
NM_001102575
NM_052870
|
SNX18
|
sorting nexin 18
|
|
chrX_+_9431314
|
2.069
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr12_-_53614121
|
2.065
|
NM_001042728
NM_001243732
|
RARG
|
retinoic acid receptor, gamma
|
|
chr1_+_64088886
|
2.056
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr16_-_87525317
|
2.038
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr13_+_28712379
|
2.022
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr1_+_64059479
|
2.016
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr10_-_65225605
|
2.015
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr10_-_61666236
|
2.011
|
NM_005436
|
CCDC6
|
coiled-coil domain containing 6
|
|
chr19_+_54695103
|
2.005
|
NM_001077446
|
TSEN34
|
tRNA splicing endonuclease 34 homolog (S. cerevisiae)
|
|
chr17_-_48474910
|
2.001
|
NM_018509
|
LRRC59
|
leucine rich repeat containing 59
|
|
chr5_-_88199868
|
1.988
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr19_+_56187990
|
1.988
|
NM_001130071
|
EPN1
|
epsin 1
|
|
chr14_-_31676680
|
1.925
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr3_-_176915012
|
1.923
|
NM_024665
|
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1
|
|
chr10_+_98592015
|
1.890
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr17_+_73521744
|
1.884
|
NM_001015002
NM_001031803
NM_004524
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr16_+_67143783
|
1.862
|
NM_025187
|
C16orf70
|
chromosome 16 open reading frame 70
|
|
chr4_-_165304406
|
1.840
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr3_+_63850181
|
1.819
|
NM_000333
|
ATXN7
|
ataxin 7
|
|
chr7_+_21467572
|
1.819
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr5_-_88119604
|
1.814
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_+_149639435
|
1.794
|
NM_015093
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2
|
|
chr1_+_87170222
|
1.758
|
NM_001206651
NM_001206652
NM_001206653
NM_016009
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1
|
|
chr10_+_11047253
|
1.756
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr11_+_120207617
|
1.754
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr6_-_134639195
|
1.742
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_-_65357249
|
1.740
|
NM_004161
NM_015543
|
RAB1A
|
RAB1A, member RAS oncogene family
|
|
chr17_-_48785016
|
1.730
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr4_+_113066552
|
1.692
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr2_+_198380294
|
1.692
|
NM_199482
|
MOB4
|
MOB family member 4, phocein
|
|
chrY_-_15592549
|
1.682
|
NM_007125
NM_182659
NM_182660
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat gene, Y-linked
|
|
chr17_-_8770976
|
1.663
|
NM_001010855
|
PIK3R6
|
phosphoinositide-3-kinase, regulatory subunit 6
|
|
chr19_-_11450254
|
1.643
|
NM_004283
|
RAB3D
|
RAB3D, member RAS oncogene family
|
|
chr1_-_204380903
|
1.643
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr22_+_37678423
|
1.641
|
NM_013385
|
CYTH4
|
cytohesin 4
|
|
chr11_+_13299260
|
1.639
|
NM_001030272
NM_001030273
NM_001178
|
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr3_+_187943192
|
1.638
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr1_-_31538553
|
1.637
|
NM_001020658
NM_014676
|
PUM1
|
pumilio homolog 1 (Drosophila)
|
|
chr8_-_116681103
|
1.626
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr1_-_184724021
|
1.612
|
NM_025191
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3
|
|
chr6_-_30712294
|
1.602
|
NM_003897
|
IER3
|
immediate early response 3
|
|
chr19_-_15560761
|
1.588
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr6_-_31628509
|
1.583
|
NM_021184
|
C6orf47
|
chromosome 6 open reading frame 47
|
|
chr8_+_81398416
|
1.566
|
NM_001105539
NM_023929
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr13_-_31040035
|
1.534
|
NM_002128
|
HMGB1
|
high mobility group box 1
|
|
chr13_+_49549953
|
1.530
|
NM_001079673
|
FNDC3A
|
fibronectin type III domain containing 3A
|
|
chr19_+_16178170
|
1.526
|
NM_001145160
|
TPM4
|
tropomyosin 4
|
|
chr3_+_20081523
|
1.460
|
NM_003884
|
KAT2B
|
K(lysine) acetyltransferase 2B
|
|
chrX_+_44732402
|
1.444
|
NM_021140
|
KDM6A
|
lysine (K)-specific demethylase 6A
|
|
chr6_-_74363689
|
1.441
|
NM_012434
|
SLC17A5
|
solute carrier family 17 (anion/sugar transporter), member 5
|
|
chr10_-_11653529
|
1.428
|
NM_014688
|
USP6NL
|
USP6 N-terminal like
|
|
chr1_+_24069855
|
1.426
|
NM_003198
|
TCEB3
|
transcription elongation factor B (SIII), polypeptide 3 (110kDa, elongin A)
|
|
chr20_-_5591487
|
1.426
|
NM_019593
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae)
|
|
chr3_+_187871473
|
1.425
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_-_11574273
|
1.422
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr2_-_61765394
|
1.394
|
NM_003400
|
XPO1
|
exportin 1 (CRM1 homolog, yeast)
|
|
chr17_+_7210841
|
1.390
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr5_+_42550181
|
1.389
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr5_-_55290601
|
1.370
|
NM_001190981
NM_002184
NM_175767
|
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor)
|
|
chr3_-_52931595
|
1.334
|
NM_001198974
NM_198563
|
TMEM110-MUSTN1
TMEM110
|
TMEM110-MUSTN1 readthrough
transmembrane protein 110
|
|
chr9_-_80646150
|
1.327
|
NM_002072
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide
|
|
chr5_+_42548314
|
1.322
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr3_-_112218383
|
1.310
|
NM_001085357
NM_181780
|
BTLA
|
B and T lymphocyte associated
|
|
chr1_+_64058891
|
1.309
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr6_+_34204647
|
1.306
|
NM_145901
NM_145902
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr5_+_42549657
|
1.304
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr7_+_28725597
|
1.296
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_-_142783225
|
1.292
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr6_+_34204576
|
1.289
|
NM_002131
NM_145899
NM_145903
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr1_-_10856704
|
1.265
|
NM_001079843
NM_017766
|
CASZ1
|
castor zinc finger 1
|
|
chr22_+_46546498
|
1.260
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr14_+_52781004
|
1.259
|
NM_000956
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa
|
|
chr7_+_100271309
|
1.256
|
NM_005273
|
GNB2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2
|
|
chr7_+_28452143
|
1.241
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_+_42565562
|
1.219
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr5_+_42548144
|
1.213
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr5_+_6746100
|
1.211
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr6_-_134497069
|
1.204
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr1_+_117910052
|
1.199
|
NM_006699
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chr10_+_63661012
|
1.196
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr11_+_118307072
|
1.186
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr3_+_122513870
|
1.185
|
NM_032839
|
DIRC2
|
disrupted in renal carcinoma 2
|
|
chr2_-_64371444
|
1.161
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chrX_-_41782230
|
1.146
|
NM_001126054
NM_001126055
NM_003688
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family)
|
|
chr6_+_152128685
|
1.142
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr12_-_48298765
|
1.140
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr12_+_65004191
|
1.123
|
NM_178169
|
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr20_-_30310597
|
1.121
|
NM_001191
NM_138578
|
BCL2L1
|
BCL2-like 1
|
|
chr11_-_8680382
|
1.113
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr8_-_28243941
|
1.099
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr9_+_5450455
|
1.098
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr1_+_156163722
|
1.092
|
NM_014655
|
SLC25A44
|
solute carrier family 25, member 44
|
|
chr11_+_126152979
|
1.089
|
NM_001039661
NM_148910
|
TIRAP
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein
|
|
chr9_+_116638544
|
1.076
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr17_+_7211260
|
1.075
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr11_+_107879407
|
1.075
|
NM_003478
|
CUL5
|
cullin 5
|
|
chr6_+_152126807
|
1.070
|
NM_001122741
|
ESR1
|
estrogen receptor 1
|
|
chr5_-_88179278
|
1.054
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_-_53499055
|
1.043
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr17_+_7210317
|
1.037
|
NM_001143760
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr6_-_134496006
|
1.033
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr22_-_31742217
|
1.033
|
NM_014323
NM_032050
NM_032051
NM_032052
|
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1
|
|
chr7_+_28475233
|
1.032
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr22_-_42322778
|
1.032
|
NM_052945
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C
|
|
chr11_-_85780899
|
1.029
|
NM_001206947
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr9_-_115095933
|
1.027
|
NM_001163788
NM_001163790
NM_001244896
NM_001244897
NM_005156
|
PTBP3
|
polypyrimidine tract binding protein 3
|
|
chr6_+_152011630
|
1.026
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr5_+_42423811
|
1.011
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr5_+_42425093
|
1.007
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr5_+_42565963
|
0.995
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr14_+_65453439
|
0.993
|
NM_002028
|
FNTB
|
farnesyltransferase, CAAX box, beta
|
|
chr6_-_134498973
|
0.993
|
NM_001143677
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr9_-_110252045
|
0.991
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr1_+_114493766
|
0.986
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr20_+_49348050
|
0.976
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr1_-_179198814
|
0.962
|
NM_001136001
NM_001168236
NM_001168237
NM_001168238
NM_007314
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr6_-_132834165
|
0.915
|
NM_003569
|
STX7
|
syntaxin 7
|
|
chr6_-_88411984
|
0.913
|
NM_018064
|
AKIRIN2
|
akirin 2
|
|
chr9_+_91606349
|
0.912
|
NM_005226
|
S1PR3
|
sphingosine-1-phosphate receptor 3
|
|
chr16_-_18937722
|
0.911
|
NM_015092
|
SMG1
|
smg-1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr5_+_42424553
|
0.889
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr1_+_101361589
|
0.875
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|