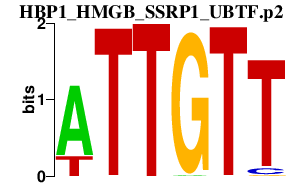
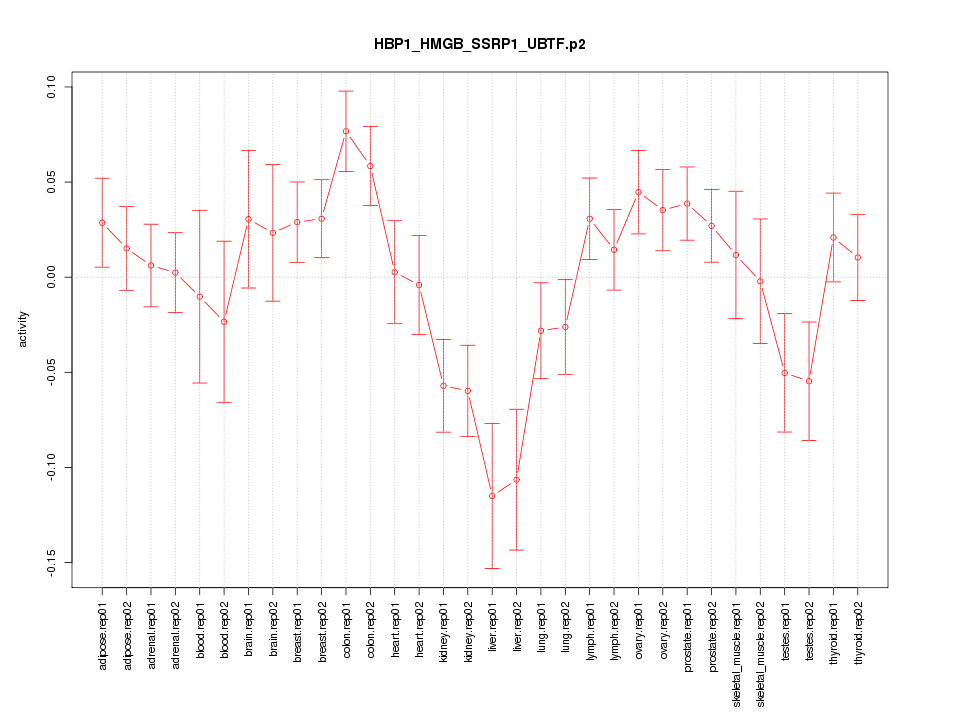
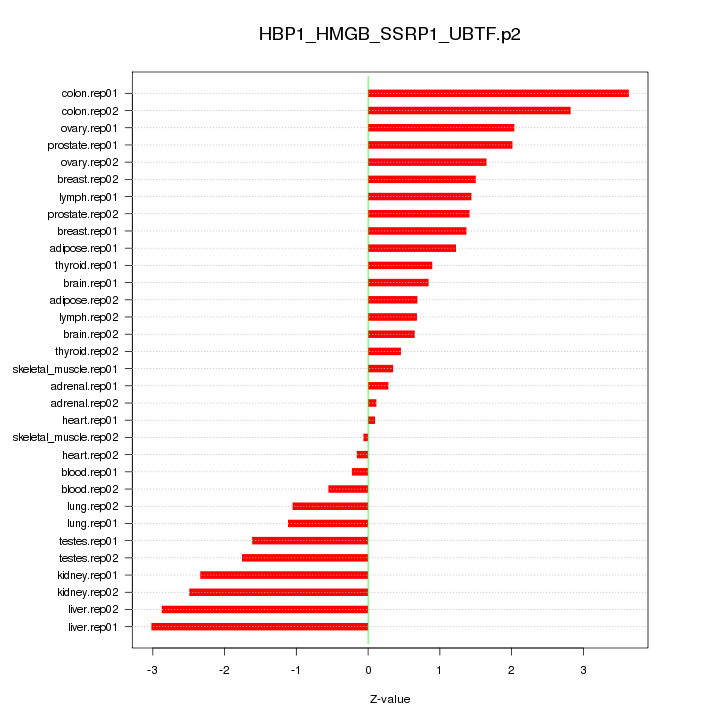
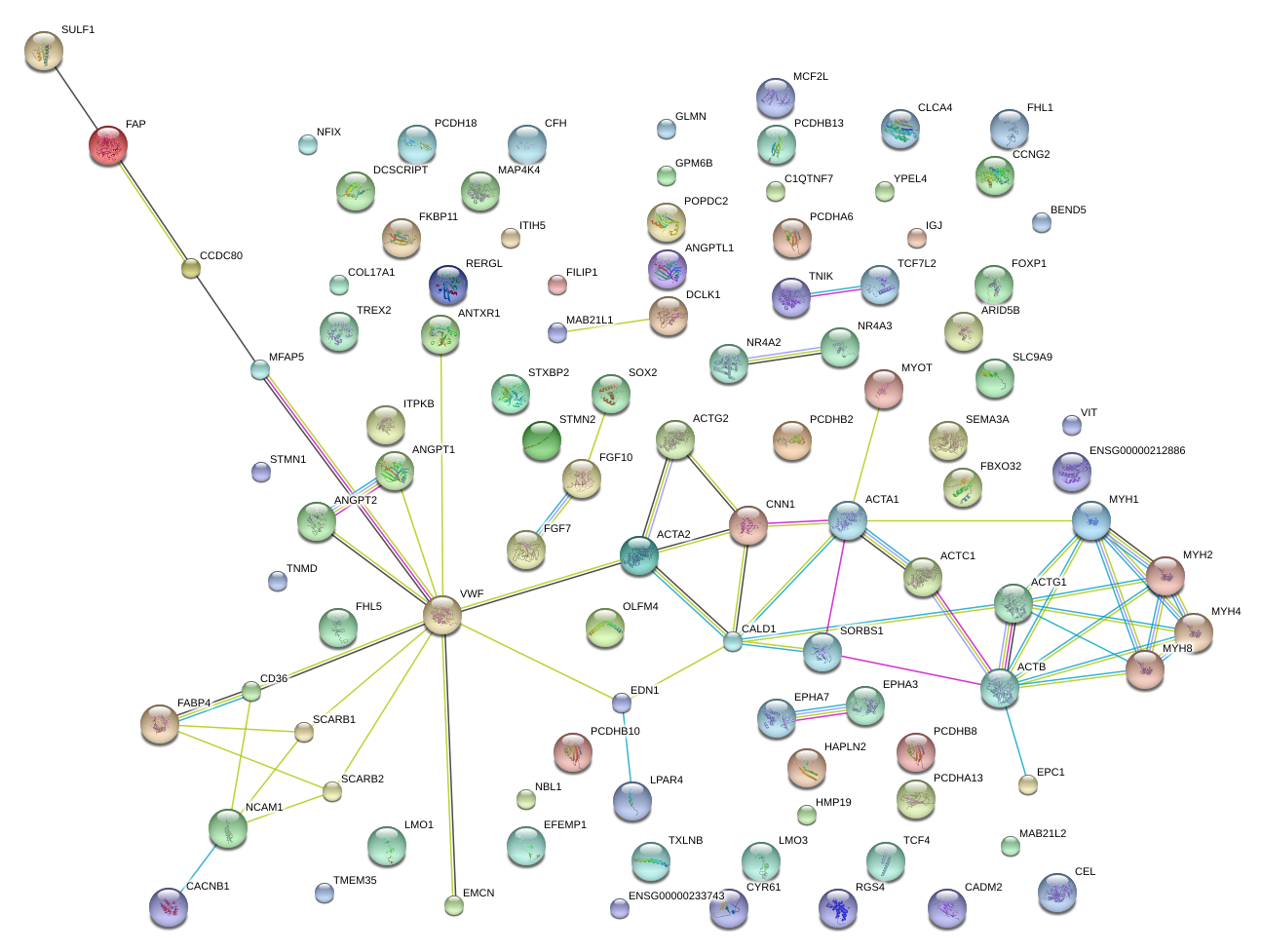
|
chr7_-_83824216
|
6.534
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr11_+_112831968
|
6.182
|
NM_000615
NM_001076682
NM_001242607
NM_001242608
NM_181351
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr8_-_82395401
|
6.003
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr15_+_49715374
|
5.662
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr8_-_108510094
|
5.308
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr6_-_139613114
|
5.100
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr8_-_124553448
|
4.986
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr11_+_112832089
|
4.797
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr8_-_124553253
|
4.762
|
|
FBXO32
|
F-box protein 32
|
|
chrX_+_100333835
|
4.507
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr5_-_44388666
|
4.292
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chrX_+_99839789
|
4.069
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr12_-_16757944
|
3.963
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_+_135251454
|
3.800
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr18_-_53071225
|
3.510
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr12_-_16759531
|
3.461
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_+_13134782
|
3.362
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr12_-_16760935
|
3.359
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_16759430
|
3.346
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_-_8815266
|
3.324
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr3_+_181417387
|
3.313
|
|
SOX2-OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr11_+_112832281
|
3.215
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr12_-_16758312
|
3.173
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_69240137
|
3.126
|
NM_018153
NM_032208
NM_053034
|
ANTXR1
|
anthrax toxin receptor 1
|
|
chr12_-_16759420
|
3.048
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_102508333
|
2.961
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr18_-_52988916
|
2.915
|
NM_001243234
NM_001243235
|
TCF4
|
transcription factor 4
|
|
chr3_+_89156804
|
2.883
|
|
EPHA3
|
EPH receptor A3
|
|
chr13_-_36429798
|
2.850
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr18_-_53177999
|
2.843
|
NM_001243231
|
TCF4
|
transcription factor 4
|
|
chr5_+_137203559
|
2.824
|
|
MYOT
|
myotilin
|
|
chr4_-_101439071
|
2.809
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr4_+_15429628
|
2.798
|
NM_031911
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chrX_-_13835213
|
2.793
|
|
GPM6B
|
glycoprotein M6B
|
|
chr10_+_63661012
|
2.778
|
NM_032199
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr4_-_138453628
|
2.772
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr12_-_8815370
|
2.754
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chrX_+_135251795
|
2.722
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr7_+_80267782
|
2.692
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr8_+_70405080
|
2.690
|
|
SULF1
|
sulfatase 1
|
|
chrX_-_13835012
|
2.663
|
|
GPM6B
|
glycoprotein M6B
|
|
chr12_-_16758839
|
2.637
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_-_13835187
|
2.622
|
|
GPM6B
|
glycoprotein M6B
|
|
chr6_+_97010423
|
2.607
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr11_+_112832201
|
2.594
|
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr3_-_119379269
|
2.565
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr12_-_18243052
|
2.557
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr5_+_140557370
|
2.472
|
NM_019120
|
PCDHB8
|
protocadherin beta 8
|
|
chr13_+_113633625
|
2.441
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr6_+_12290528
|
2.432
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr2_+_74120132
|
2.425
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr4_-_71532341
|
2.415
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chrX_+_78003205
|
2.409
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chrX_-_13835313
|
2.372
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr4_+_151503076
|
2.370
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr5_+_140571903
|
2.368
|
NM_018930
|
PCDHB10
|
protocadherin beta 10
|
|
chr6_-_76203256
|
2.344
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr5_+_137203544
|
2.308
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr2_-_157189040
|
2.307
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_-_226926875
|
2.299
|
NM_002221
|
ITPKB
|
inositol-trisphosphate 3-kinase B
|
|
chr1_+_163038564
|
2.281
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr2_+_74120092
|
2.277
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr7_+_80267922
|
2.258
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr3_+_85775631
|
2.256
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr1_-_26233367
|
2.255
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr3_+_181429711
|
2.247
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr19_+_13135745
|
2.244
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr8_-_6420454
|
2.239
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr19_+_13135809
|
2.238
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr17_-_10452939
|
2.211
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr5_-_71803119
|
2.188
|
NM_152625
|
ZNF366
|
zinc finger protein 366
|
|
chr10_-_32667543
|
2.149
|
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr1_+_86046434
|
2.143
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr3_-_112359443
|
2.087
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr18_-_53257033
|
2.077
|
NM_001243227
NM_001243228
|
TCF4
|
transcription factor 4
|
|
chr4_-_71532206
|
2.076
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr2_-_56412904
|
2.060
|
|
LOC100129434
|
uncharacterized LOC100129434
|
|
chr5_-_87969135
|
2.056
|
|
LINC00461
|
long intergenic non-protein coding RNA 461
|
|
chr8_+_70405027
|
2.045
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr2_+_36923832
|
2.041
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr13_+_53602829
|
2.039
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr2_-_163099877
|
2.021
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr5_+_173472692
|
2.005
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr1_-_178840079
|
1.994
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr3_+_89156673
|
1.990
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr5_+_140474198
|
1.974
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr7_+_134464372
|
1.961
|
|
CALD1
|
caldesmon 1
|
|
chr10_-_7682741
|
1.953
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr1_-_49242406
|
1.951
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr2_-_56151273
|
1.948
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr3_-_71353910
|
1.944
|
NM_001244812
|
FOXP1
|
forkhead box P1
|
|
chr11_-_57417247
|
1.924
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_-_112359547
|
1.914
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr10_-_105845619
|
1.897
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr4_+_78078394
|
1.886
|
|
CCNG2
|
cyclin G2
|
|
chr1_+_156589085
|
1.883
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr6_-_94129059
|
1.873
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr10_-_97200895
|
1.870
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr1_+_87012758
|
1.855
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr12_-_6233684
|
1.852
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr8_+_80523048
|
1.832
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|
|
chr1_+_19972259
|
1.821
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr5_+_140235594
|
1.820
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr19_+_11649578
|
1.808
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr3_-_143567163
|
1.795
|
NM_173653
|
SLC9A9
|
solute carrier family 9 (sodium/hydrogen exchanger), member 9
|
|
chr3_+_173116243
|
1.795
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr19_+_13135368
|
1.787
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr1_-_201476218
|
1.783
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr3_-_112359565
|
1.781
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_19157262
|
1.768
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr1_+_155853546
|
1.763
|
|
SYT11
|
synaptotagmin XI
|
|
chr3_+_112931374
|
1.755
|
NM_033254
|
BOC
|
Boc homolog (mouse)
|
|
chr2_+_169923531
|
1.747
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr5_-_83680590
|
1.742
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr6_-_56716412
|
1.734
|
|
DST
|
dystonin
|
|
chr5_+_15500288
|
1.727
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr5_+_173472737
|
1.724
|
|
HMP19
|
HMP19 protein
|
|
chr8_+_80523351
|
1.710
|
|
STMN2
|
stathmin-like 2
|
|
chr4_+_183245136
|
1.709
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr12_-_16759939
|
1.705
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_83680191
|
1.698
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr9_+_75766777
|
1.692
|
NM_000700
|
ANXA1
|
annexin A1
|
|
chr7_-_7575449
|
1.683
|
NM_001037763
|
COL28A1
|
collagen, type XXVIII, alpha 1
|
|
chr2_-_224467049
|
1.662
|
|
SCG2
|
secretogranin II
|
|
chr21_-_36231794
|
1.635
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr12_-_8815479
|
1.630
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr18_-_45663667
|
1.628
|
NM_001039360
|
ZBTB7C
|
zinc finger and BTB domain containing 7C
|
|
chr7_+_114066554
|
1.626
|
NM_001172767
NM_148899
|
FOXP2
|
forkhead box P2
|
|
chr4_-_111558150
|
1.621
|
|
PITX2
|
paired-like homeodomain 2
|
|
chrX_-_128788913
|
1.620
|
NM_017413
|
APLN
|
apelin
|
|
chr1_-_244006885
|
1.612
|
NM_001206729
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma)
|
|
chr17_-_67240955
|
1.601
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr6_-_123957941
|
1.601
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr11_-_101787252
|
1.595
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr5_-_146833150
|
1.593
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr8_-_93075190
|
1.592
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_94423766
|
1.590
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr6_-_34524083
|
1.583
|
NM_001252294
NM_012391
|
SPDEF
|
SAM pointed domain containing ets transcription factor
|
|
chr12_-_28124915
|
1.569
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr1_+_228337414
|
1.561
|
NM_020435
|
GJC2
|
gap junction protein, gamma 2, 47kDa
|
|
chr1_-_201475966
|
1.560
|
NM_001193571
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr2_+_102508377
|
1.560
|
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr10_+_86184685
|
1.558
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr20_+_17207680
|
1.547
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr16_-_15835382
|
1.546
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr11_+_36317724
|
1.546
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr2_-_163008913
|
1.546
|
NM_002054
|
GCG
|
glucagon
|
|
chr6_-_29644930
|
1.543
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr10_-_104179468
|
1.543
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr8_-_93029864
|
1.527
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_+_15500736
|
1.523
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chrX_+_15518899
|
1.521
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr18_+_3594056
|
1.520
|
|
FLJ35776
|
uncharacterized LOC649446
|
|
chr12_-_16759733
|
1.517
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_192635627
|
1.509
|
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr12_+_49212253
|
1.506
|
NM_000725
NM_001206915
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr15_-_83876769
|
1.501
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr8_+_80523376
|
1.491
|
|
STMN2
|
stathmin-like 2
|
|
chr4_+_119809978
|
1.486
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr17_-_39041436
|
1.485
|
NM_019010
|
KRT20
|
keratin 20
|
|
chr1_-_201476248
|
1.482
|
NM_001193570
NM_004078
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr11_+_131780537
|
1.454
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr1_-_201476241
|
1.454
|
|
CSRP1
|
cysteine and glycine-rich protein 1
|
|
chr5_+_140588267
|
1.438
|
NM_018932
|
PCDHB12
|
protocadherin beta 12
|
|
chr3_-_195306274
|
1.431
|
|
APOD
|
apolipoprotein D
|
|
chrX_+_107683015
|
1.430
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr4_-_88450243
|
1.430
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr7_+_134576187
|
1.429
|
|
CALD1
|
caldesmon 1
|
|
chr11_-_111782447
|
1.426
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr17_-_66951381
|
1.416
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr1_+_13911966
|
1.415
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr12_+_13349716
|
1.412
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr14_+_23305792
|
1.410
|
NM_004995
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr12_-_40013842
|
1.406
|
NM_005164
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr14_+_85996512
|
1.404
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr7_-_16921596
|
1.395
|
NM_176813
|
AGR3
|
anterior gradient 3 homolog (Xenopus laevis)
|
|
chr5_+_15500539
|
1.394
|
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr10_-_116251582
|
1.385
|
|
|
|
|
chr7_+_94292816
|
1.385
|
|
PEG10
|
paternally expressed 10
|
|
chr7_-_151107117
|
1.384
|
NM_198285
|
WDR86
|
WD repeat domain 86
|
|
chr18_-_40857245
|
1.382
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr3_+_115342302
|
1.382
|
|
GAP43
|
growth associated protein 43
|
|
chr14_-_60097531
|
1.379
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr9_-_14313509
|
1.377
|
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_118869441
|
1.374
|
NM_002667
|
PLN
|
phospholamban
|
|
chr7_+_129015483
|
1.362
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chrX_-_107682600
|
1.360
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr18_-_29993195
|
1.352
|
|
|
|
|
chr12_-_91573264
|
1.351
|
NM_133503
|
DCN
|
decorin
|
|
chr17_-_67057046
|
1.348
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr7_+_80267972
|
1.347
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr2_-_224467095
|
1.341
|
|
SCG2
|
secretogranin II
|
|
chr3_-_167191808
|
1.340
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr15_-_83876165
|
1.338
|
NM_016073
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr5_+_148786439
|
1.330
|
|
MIR143HG
|
MIR143 host gene (non-protein coding)
|
|
chr17_+_19990334
|
1.329
|
NM_001033553
NM_152904
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr20_+_17207596
|
1.328
|
NM_001201529
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_-_71542628
|
1.327
|
|
FOXP1
|
forkhead box P1
|
|
chr9_+_125795787
|
1.326
|
|
RABGAP1
GPR21
|
RAB GTPase activating protein 1
G protein-coupled receptor 21
|
|
chr1_+_84768333
|
1.325
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|