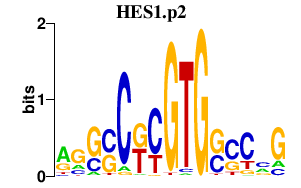
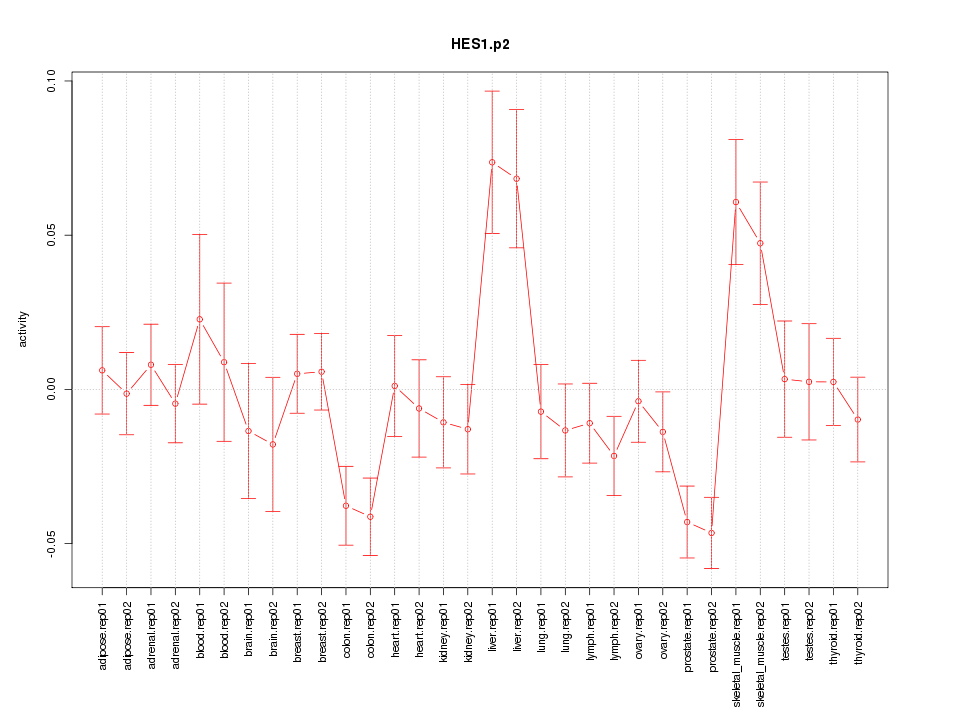
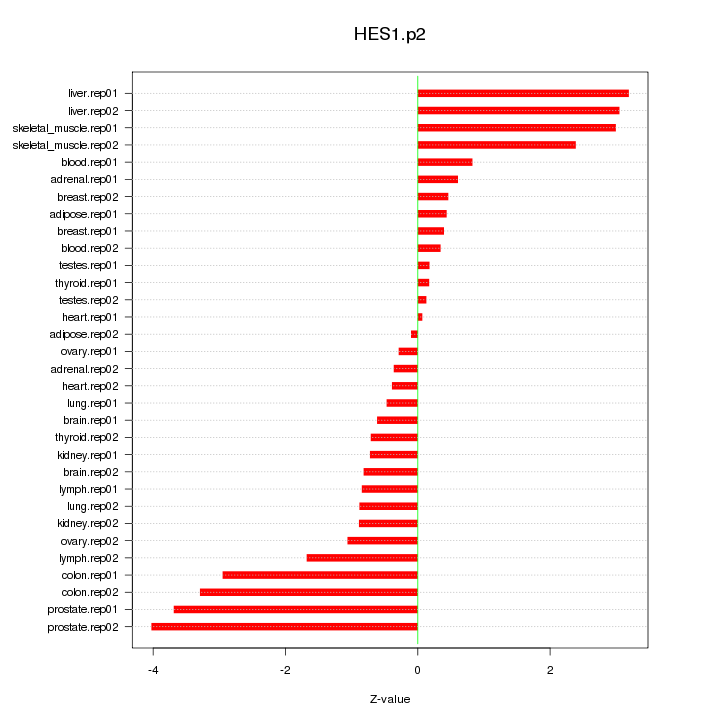
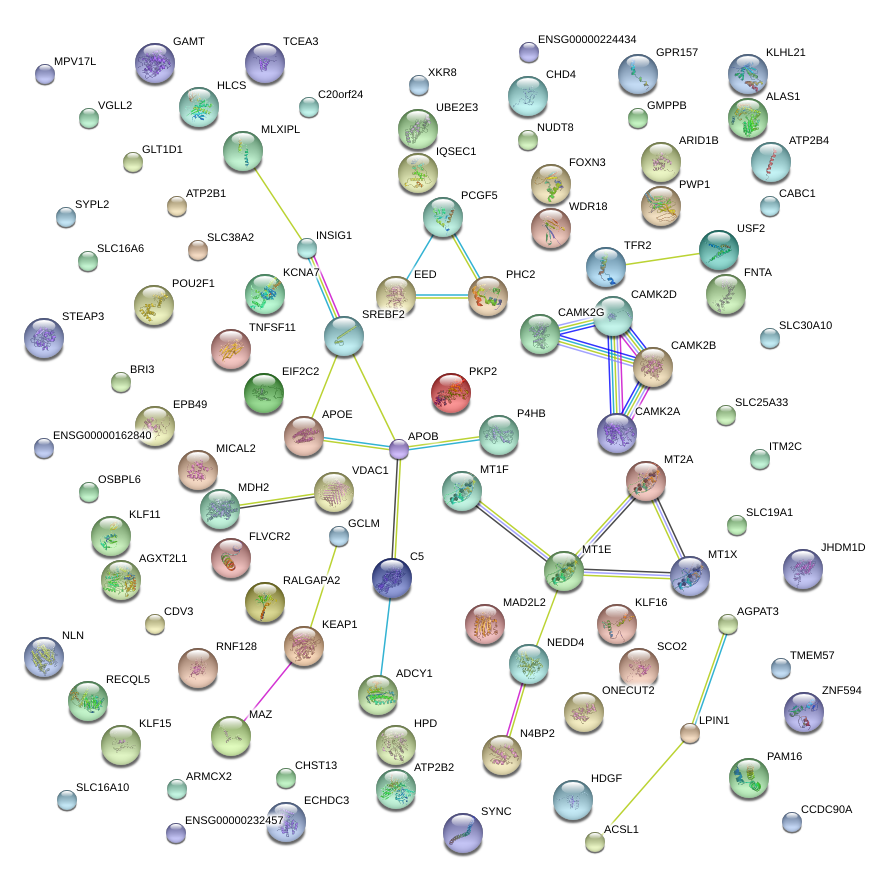
|
chr3_+_126261178
|
3.913
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr7_+_155089614
|
2.995
|
|
INSIG1
|
insulin induced gene 1
|
|
chr4_-_185747051
|
2.641
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr7_-_100230676
|
2.316
|
|
TFR2
|
transferrin receptor 2
|
|
chr6_+_111408780
|
2.088
|
NM_018593
|
SLC16A10
|
solute carrier family 16, member 10 (aromatic amino acid transporter)
|
|
chr19_-_1401474
|
2.026
|
NM_000156
NM_138924
|
GAMT
|
guanidinoacetate N-methyltransferase
|
|
chr10_+_11784355
|
1.994
|
NM_024693
|
ECHDC3
|
enoyl CoA hydratase domain containing 3
|
|
chr7_+_155089483
|
1.965
|
NM_005542
NM_198336
NM_198337
|
INSIG1
|
insulin induced gene 1
|
|
chr7_-_44365019
|
1.897
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr16_+_29817810
|
1.892
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr16_+_15489586
|
1.815
|
NM_001128423
NM_173803
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr2_-_21266915
|
1.804
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr17_-_4693595
|
1.737
|
|
LOC284014
|
uncharacterized LOC284014
|
|
chr20_+_35234210
|
1.722
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr14_-_89883306
|
1.704
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr1_-_94374986
|
1.696
|
NM_002061
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr1_-_9189087
|
1.682
|
NM_024980
|
GPR157
|
G protein-coupled receptor 157
|
|
chr11_-_67397383
|
1.678
|
NM_001243750
NM_181843
|
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8
|
|
chr2_-_200715842
|
1.669
|
|
FONG
|
uncharacterized LOC348751
|
|
chr2_+_119981383
|
1.663
|
NM_001008410
NM_018234
NM_182915
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr16_+_56642477
|
1.639
|
NM_005953
|
MT2A
|
metallothionein 2A
|
|
chr2_+_119981389
|
1.628
|
|
STEAP3
|
STEAP family member 3, metalloreductase
|
|
chr1_-_94374949
|
1.625
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr15_-_56285732
|
1.531
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr21_-_46962281
|
1.529
|
NM_001205206
NM_194255
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1
|
|
chr7_+_45614081
|
1.490
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr7_-_73038814
|
1.477
|
NM_032951
NM_032952
NM_032953
NM_032954
|
MLXIPL
|
MLX interacting protein-like
|
|
chr1_-_156721501
|
1.469
|
NM_004494
|
HDGF
|
hepatoma-derived growth factor
|
|
chr19_-_49575878
|
1.458
|
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr1_+_25757381
|
1.431
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr1_-_94375028
|
1.423
|
|
GCLM
|
glutamate-cysteine ligase, modifier subunit
|
|
chr10_+_92922696
|
1.397
|
|
PCGF5
|
polycomb group ring finger 5
|
|
chr14_+_76044939
|
1.391
|
NM_017791
|
FLVCR2
|
feline leukemia virus subgroup C cellular receptor family, member 2
|
|
chr2_+_181845605
|
1.375
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr1_-_220101925
|
1.371
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr16_+_15489619
|
1.346
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr20_-_20693122
|
1.299
|
NM_020343
|
RALGAPA2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic)
|
|
chr2_-_200715895
|
1.289
|
|
FONG
|
uncharacterized LOC348751
|
|
chr19_+_35759880
|
1.287
|
NM_003367
NM_207291
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr12_+_129338051
|
1.286
|
NM_144669
|
GLT1D1
|
glycosyltransferase 1 domain containing 1
|
|
chr6_+_157099041
|
1.281
|
NM_017519
NM_020732
|
ARID1B
|
AT rich interactive domain 1B (SWI1-like)
|
|
chr1_+_110009099
|
1.266
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr9_-_123837410
|
1.243
|
|
C5
|
complement component 5
|
|
chr22_-_50964573
|
1.241
|
NM_001169110
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr1_-_23751126
|
1.240
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr20_+_35234152
|
1.235
|
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr1_+_110009220
|
1.218
|
|
SYPL2
|
synaptophysin-like 2
|
|
chr18_+_77794345
|
1.207
|
NM_001171967
NM_024805
|
RBFA
|
ribosome binding factor A (putative)
|
|
chr12_-_33049664
|
1.202
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr20_+_35234077
|
1.187
|
NM_001199534
NM_018840
NM_199483
|
C20orf24
|
chromosome 20 open reading frame 24
|
|
chr17_-_79818370
|
1.185
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr8_-_141645603
|
1.182
|
NM_001164623
NM_012154
|
EIF2C2
|
eukaryotic translation initiation factor 2C, 2
|
|
chr19_-_10613767
|
1.162
|
NM_203500
|
KEAP1
|
kelch-like ECH-associated protein 1
|
|
chr3_-_10547267
|
1.158
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr2_+_11886739
|
1.150
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr19_-_1863425
|
1.137
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr13_+_43148182
|
1.135
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr5_-_133340823
|
1.130
|
NM_003374
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr1_-_33815411
|
1.115
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr5_+_65017993
|
1.114
|
NM_020726
|
NLN
|
neurolysin (metallopeptidase M3 family)
|
|
chr4_-_109684053
|
1.107
|
NM_001146590
NM_001146627
NM_031279
|
AGXT2L1
|
alanine-glyoxylate aminotransferase 2-like 1
|
|
chr16_-_4401265
|
1.097
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr1_+_167189948
|
1.092
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr17_-_79818418
|
1.090
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr1_+_9599546
|
1.085
|
|
SLC25A33
|
solute carrier family 25, member 33
|
|
chr7_-_139876718
|
1.080
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr4_+_40058507
|
1.078
|
NM_018177
|
N4BP2
|
NEDD4 binding protein 2
|
|
chr19_+_35759967
|
1.072
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chrX_+_105969875
|
1.070
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr1_-_6662681
|
1.067
|
|
KLHL21
|
kelch-like 21 (Drosophila)
|
|
chr17_-_66287256
|
1.047
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr7_+_75677356
|
1.042
|
NM_005918
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial)
|
|
chr17_-_79818444
|
1.031
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr19_+_984316
|
1.022
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr2_+_181845088
|
1.020
|
NM_182678
NM_006357
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr3_-_126076057
|
1.016
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr12_-_122326516
|
1.006
|
NM_001171993
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr2_+_10184371
|
1.005
|
NM_001177718
|
KLF11
|
Kruppel-like factor 11
|
|
chr3_+_133292557
|
0.999
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr21_+_45285026
|
0.997
|
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr6_-_13814546
|
0.993
|
NM_001031713
|
CCDC90A
|
coiled-coil domain containing 90A
|
|
chr1_+_227127937
|
0.989
|
NM_020247
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr1_-_33815340
|
0.989
|
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr1_+_227127959
|
0.969
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr7_+_75677402
|
0.967
|
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial)
|
|
chr7_-_100231135
|
0.966
|
NM_001206855
|
TFR2
|
transferrin receptor 2
|
|
chr8_+_121823538
|
0.960
|
|
|
|
|
chr2_+_181845803
|
0.955
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr21_-_38362481
|
0.954
|
NM_000411
|
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase)
|
|
chr1_-_11751677
|
0.948
|
NM_001127325
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chr3_-_187871816
|
0.941
|
|
LPP-AS2
|
LPP antisense RNA 2 (non-protein coding)
|
|
chr17_-_79818318
|
0.937
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr19_+_35760002
|
0.935
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr1_-_178007057
|
0.934
|
|
LOC730102
|
quinone oxidoreductase-like protein 2 pseudogene
|
|
chr3_+_52232179
|
0.929
|
|
ALAS1
|
aminolevulinate, delta-, synthase 1
|
|
chr11_+_12132063
|
0.922
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr1_+_227127993
|
0.920
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr3_-_10547340
|
0.918
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr3_+_133292714
|
0.915
|
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr12_-_6716475
|
0.915
|
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr7_+_97910978
|
0.912
|
NM_001159491
NM_015379
|
BRI3
|
brain protein I3
|
|
chr3_-_49761293
|
0.911
|
NM_013334
NM_021971
|
GMPPB
|
GDP-mannose pyrophosphorylase B
|
|
chr6_+_117586720
|
0.901
|
NM_153453
NM_182645
|
VGLL2
|
vestigial like 2 (Drosophila)
|
|
chr2_+_179059071
|
0.898
|
NM_001201480
NM_001201481
NM_001201482
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr3_-_13008959
|
0.893
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr12_-_46766019
|
0.891
|
|
SLC38A2
|
solute carrier family 38, member 2
|
|
chr1_-_33815498
|
0.884
|
NM_004427
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr21_+_45285101
|
0.884
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr22_-_50964856
|
0.876
|
NM_001169109
|
SCO2
|
SCO cytochrome oxidase deficient homolog 2 (yeast)
|
|
chr1_-_33168356
|
0.874
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr18_+_55102777
|
0.870
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr17_-_79818514
|
0.870
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr22_+_42229137
|
0.864
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr1_+_28286481
|
0.857
|
NM_018053
|
XKR8
|
XK, Kell blood group complex subunit-related family, member 8
|
|
chr16_-_18573334
|
0.855
|
NM_001004060
NM_173614
|
NOMO2
|
NODAL modulator 2
|
|
chr3_+_133292404
|
0.848
|
NM_001134422
NM_017548
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr1_+_227127999
|
0.846
|
|
ADCK3
|
aarF domain containing kinase 3
|
|
chr16_-_56459371
|
0.840
|
NM_001144
|
AMFR
|
autocrine motility factor receptor
|
|
chr16_+_84733611
|
0.837
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr19_-_2256415
|
0.836
|
NM_144616
|
JSRP1
|
junctional sarcoplasmic reticulum protein 1
|
|
chr1_+_94883929
|
0.835
|
NM_001122674
NM_002858
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr17_+_7348840
|
0.835
|
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr17_-_79519360
|
0.835
|
NM_025161
|
C17orf70
|
chromosome 17 open reading frame 70
|
|
chr2_-_38604430
|
0.833
|
NM_001135673
NM_022374
|
ATL2
|
atlastin GTPase 2
|
|
chr15_+_50474357
|
0.829
|
NM_001159629
NM_003645
|
SLC27A2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr8_+_1950304
|
0.828
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr17_+_46018888
|
0.827
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr11_-_71159371
|
0.822
|
|
DHCR7
|
7-dehydrocholesterol reductase
|
|
chr7_+_74379079
|
0.819
|
NM_001145063
|
GATSL1
GATSL2
|
GATS protein-like 1
GATS protein-like 2
|
|
chr9_+_139839697
|
0.817
|
NM_000606
|
C8G
|
complement component 8, gamma polypeptide
|
|
chr5_+_78365582
|
0.817
|
|
BHMT2
|
betaine--homocysteine S-methyltransferase 2
|
|
chr17_+_72428228
|
0.816
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr17_+_7348362
|
0.816
|
NM_000747
|
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle)
|
|
chr2_+_181845368
|
0.813
|
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3
|
|
chr1_+_36273786
|
0.811
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr16_+_29817426
|
0.809
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chrX_-_131623994
|
0.807
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_-_13711643
|
0.802
|
NM_005493
|
RANBP9
|
RAN binding protein 9
|
|
chr1_-_13840157
|
0.800
|
NM_001010847
|
LRRC38
|
leucine rich repeat containing 38
|
|
chr22_-_39152008
|
0.798
|
NM_001199579
NM_015374
|
SUN2
|
Sad1 and UNC84 domain containing 2
|
|
chr1_-_11120081
|
0.793
|
NM_003132
|
SRM
|
spermidine synthase
|
|
chr4_-_74964836
|
0.789
|
|
CXCL2
|
chemokine (C-X-C motif) ligand 2
|
|
chr20_+_5986560
|
0.785
|
NM_019095
|
CRLS1
|
cardiolipin synthase 1
|
|
chr3_+_16926451
|
0.783
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr17_+_45727274
|
0.780
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr19_+_3359138
|
0.777
|
NM_001245005
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr22_+_30116288
|
0.777
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr1_+_43148359
|
0.770
|
|
YBX1
|
Y box binding protein 1
|
|
chr7_+_24612714
|
0.764
|
NM_016447
|
MPP6
|
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6)
|
|
chr15_-_102029991
|
0.763
|
|
PCSK6
|
proprotein convertase subtilisin/kexin type 6
|
|
chr2_-_38604380
|
0.761
|
|
ATL2
|
atlastin GTPase 2
|
|
chr9_+_137218324
|
0.761
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr12_-_6716550
|
0.760
|
NM_001273
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr10_-_94333721
|
0.758
|
|
IDE
|
insulin-degrading enzyme
|
|
chr20_+_57466509
|
0.753
|
|
GNAS
|
GNAS complex locus
|
|
chr7_-_47621741
|
0.751
|
NM_022748
|
TNS3
|
tensin 3
|
|
chr22_+_29663997
|
0.751
|
NM_001163285
NM_001163286
NM_001163287
NM_005243
NM_013986
|
EWSR1
|
Ewing sarcoma breakpoint region 1
|
|
chr7_-_47621635
|
0.749
|
|
TNS3
|
tensin 3
|
|
chr8_+_41435666
|
0.747
|
NM_178819
|
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta)
|
|
chr4_+_108911042
|
0.746
|
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chrX_+_21392396
|
0.746
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr1_+_19638739
|
0.746
|
NM_001040125
NM_001040126
NM_017765
|
PQLC2
|
PQ loop repeat containing 2
|
|
chr5_+_75699003
|
0.745
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr1_+_29063443
|
0.744
|
NM_001172828
|
YTHDF2
|
YTH domain family, member 2
|
|
chr4_+_57774365
|
0.744
|
|
REST
|
RE1-silencing transcription factor
|
|
chr5_-_6633149
|
0.741
|
|
NSUN2
|
NOP2/Sun domain family, member 2
|
|
chr21_+_30671113
|
0.740
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr6_-_44095177
|
0.740
|
NM_032111
|
MRPL14
|
mitochondrial ribosomal protein L14
|
|
chr22_+_42229268
|
0.740
|
|
SREBF2
|
sterol regulatory element binding transcription factor 2
|
|
chr8_-_144623579
|
0.739
|
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr3_+_50242688
|
0.738
|
NM_006841
|
SLC38A3
|
solute carrier family 38, member 3
|
|
chr6_-_11044354
|
0.737
|
NM_017770
|
ELOVL2
|
ELOVL fatty acid elongase 2
|
|
chr3_+_98451537
|
0.736
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr11_-_111637085
|
0.735
|
NM_001177562
NM_001177563
NM_002716
NM_181699
NM_181700
|
PPP2R1B
|
protein phosphatase 2, regulatory subunit A, beta
|
|
chr1_+_61547625
|
0.734
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr19_-_40791265
|
0.731
|
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr16_+_84733554
|
0.730
|
NM_005153
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr16_+_84733596
|
0.729
|
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr11_+_747428
|
0.727
|
NM_006755
|
TALDO1
|
transaldolase 1
|
|
chr1_+_29063132
|
0.727
|
NM_001173128
NM_016258
|
YTHDF2
|
YTH domain family, member 2
|
|
chr4_+_108911008
|
0.726
|
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chr5_-_140998560
|
0.726
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr17_+_35849950
|
0.722
|
NM_007026
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr8_+_96037186
|
0.721
|
NM_152416
|
C8orf38
|
chromosome 8 open reading frame 38
|
|
chr1_-_156722023
|
0.718
|
NM_001126051
NM_001126050
|
HDGF
|
hepatoma-derived growth factor
|
|
chr5_-_78365371
|
0.718
|
NM_013391
|
DMGDH
|
dimethylglycine dehydrogenase
|
|
chr3_+_110790774
|
0.716
|
NM_001243288
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr11_+_109964086
|
0.716
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr11_+_747424
|
0.713
|
|
TALDO1
|
transaldolase 1
|
|
chr5_+_70883100
|
0.712
|
NM_022132
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta)
|
|
chr1_-_55352792
|
0.708
|
|
DHCR24
|
24-dehydrocholesterol reductase
|
|
chr6_-_105850765
|
0.707
|
|
PREP
|
prolyl endopeptidase
|
|
chr5_-_6633347
|
0.707
|
NM_001193455
NM_017755
|
NSUN2
|
NOP2/Sun domain family, member 2
|
|
chr4_-_84205940
|
0.703
|
NM_015697
|
COQ2
|
coenzyme Q2 homolog, prenyltransferase (yeast)
|
|
chr6_+_44095357
|
0.701
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr16_-_46865039
|
0.699
|
NM_001001436
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr9_-_6645564
|
0.699
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr8_-_144623611
|
0.697
|
NM_015117
|
ZC3H3
|
zinc finger CCCH-type containing 3
|
|
chr17_-_42402136
|
0.697
|
|
SLC25A39
|
solute carrier family 25, member 39
|
|
chr4_-_71705188
|
0.696
|
NM_001098477
|
GRSF1
|
G-rich RNA sequence binding factor 1
|