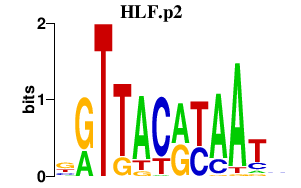
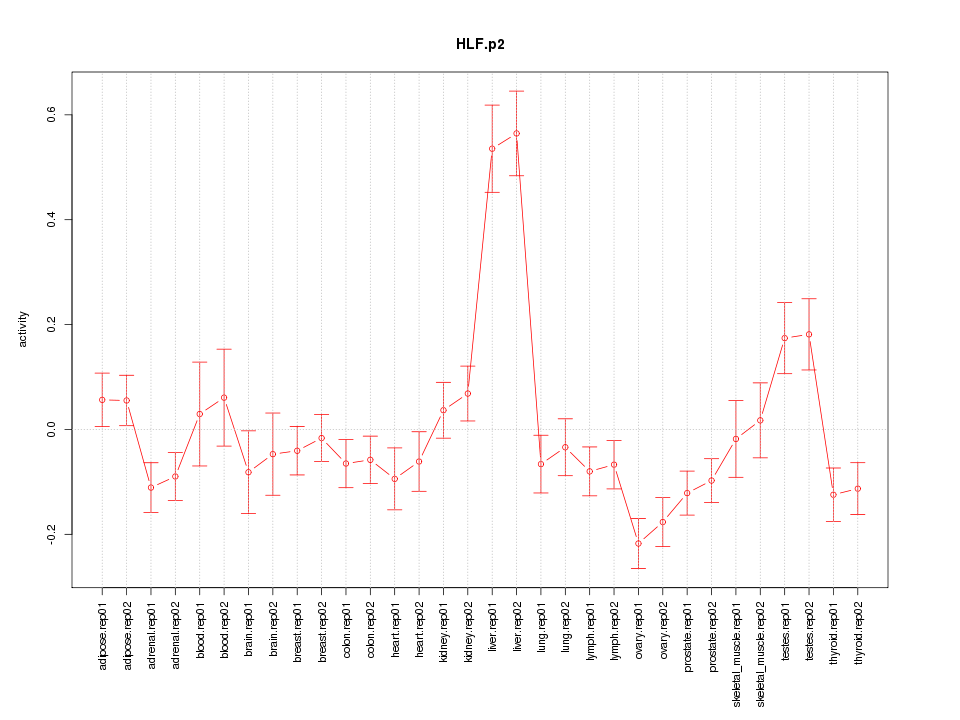
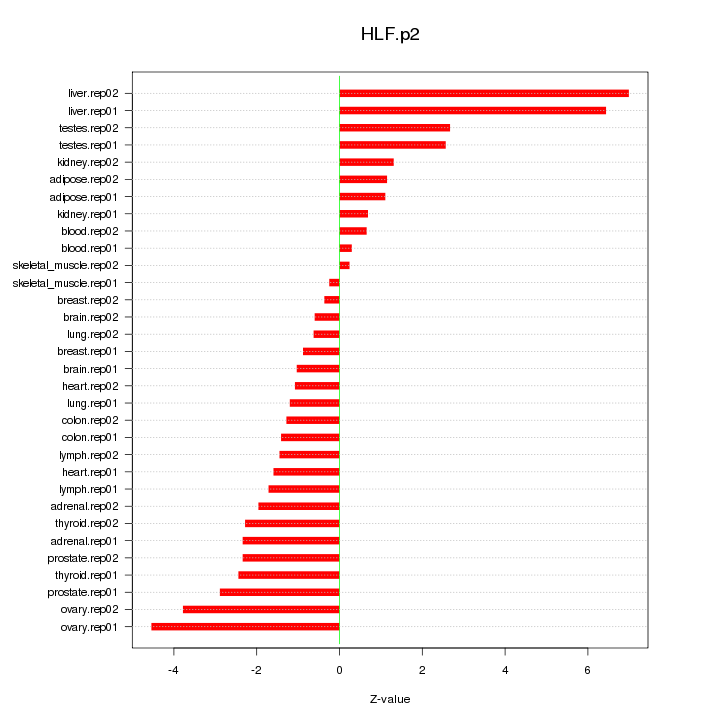
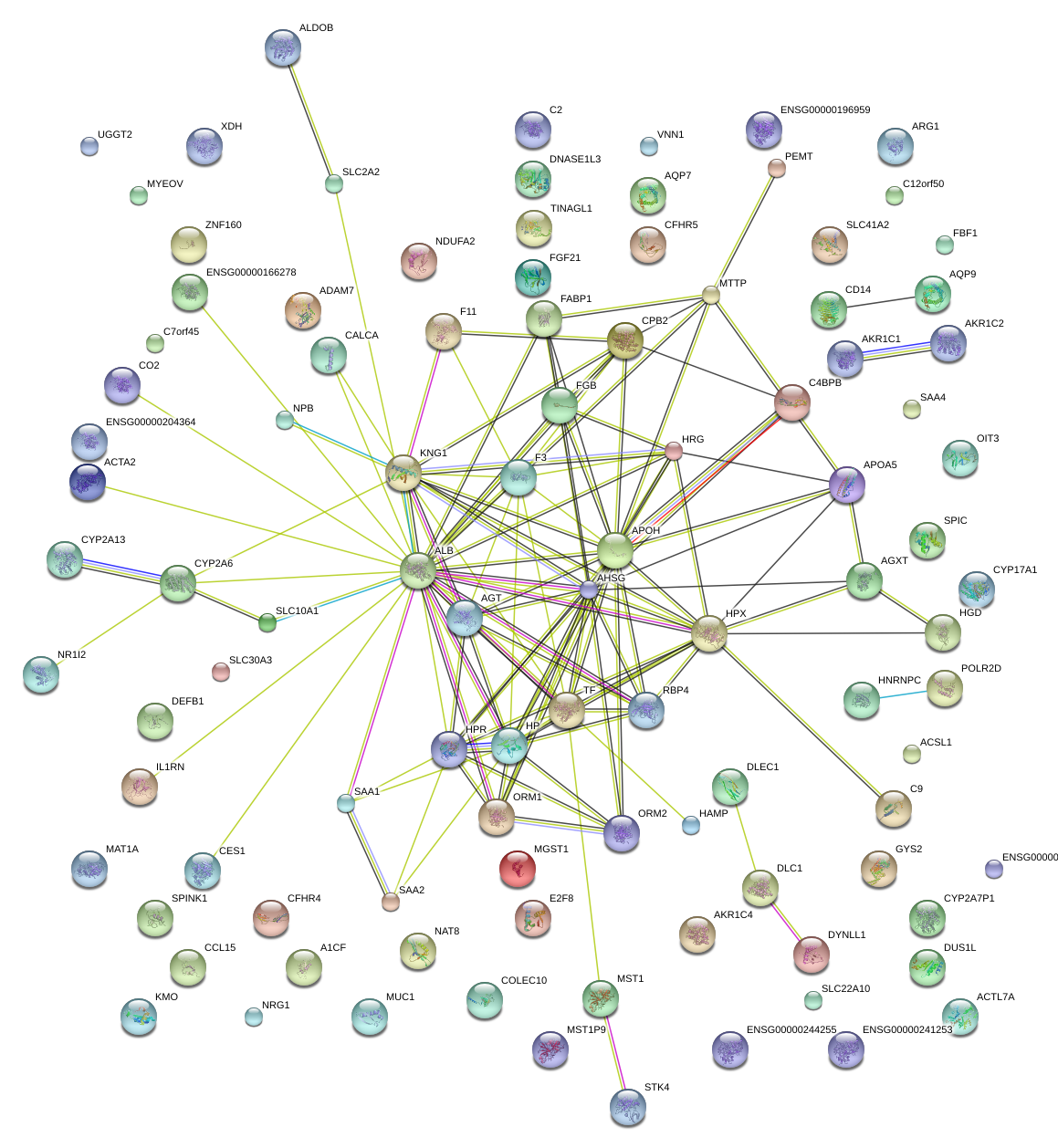
|
chr5_-_39364562
|
26.894
|
NM_001737
|
C9
|
complement component 9
|
|
chr9_+_117085302
|
22.587
|
NM_000607
|
ORM1
|
orosomucoid 1
|
|
chr3_+_186435097
|
20.122
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr9_+_117092068
|
20.062
|
NM_000608
|
ORM2
|
orosomucoid 2
|
|
chr19_-_41356193
|
17.665
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr4_+_155484131
|
17.196
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr16_+_72088507
|
16.280
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr16_+_72097124
|
14.262
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr3_+_186383770
|
14.186
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr6_+_131894343
|
14.143
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr19_+_41594367
|
13.097
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr10_-_52645430
|
13.019
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr14_-_70263871
|
12.094
|
NM_003049
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr19_+_35773409
|
11.380
|
NM_021175
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr2_-_88427535
|
10.831
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr11_-_116663106
|
10.729
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr1_+_196857143
|
10.340
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr11_+_63057371
|
10.107
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr3_+_186330844
|
10.094
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr11_-_6462129
|
9.919
|
NM_000613
|
HPX
|
hemopexin
|
|
chr11_+_18287807
|
9.349
|
NM_000331
NM_001178006
NM_199161
|
SAA1
|
serum amyloid A1
|
|
chr19_+_49259343
|
9.311
|
NM_019113
|
FGF21
|
fibroblast growth factor 21
|
|
chr10_+_74653338
|
9.216
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr3_+_119501556
|
8.894
|
NM_022002
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr12_-_21757752
|
8.863
|
NM_021957
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr11_-_14993790
|
8.442
|
NM_001033952
NM_001033953
NM_001741
|
CALCA
|
calcitonin-related polypeptide alpha
|
|
chr4_+_187187342
|
8.420
|
|
F11
|
coagulation factor XI
|
|
chr1_-_230849936
|
8.349
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_-_49726081
|
8.277
|
NM_020998
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like)
|
|
chr2_+_113885137
|
7.924
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_+_133494275
|
7.874
|
|
TF
|
transferrin
|
|
chr3_-_120400957
|
7.674
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr5_-_147211141
|
7.587
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr7_-_15601574
|
7.467
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr10_-_82049178
|
7.449
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr2_-_31637598
|
7.428
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr13_-_46679143
|
7.366
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr11_-_18258383
|
7.234
|
NM_006512
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr1_-_230849863
|
7.224
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_+_186330859
|
7.194
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr19_+_41430123
|
6.707
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr3_-_58200397
|
6.673
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr19_-_41388656
|
6.656
|
NM_000764
NM_030589
|
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7
|
|
chr2_+_113885145
|
6.612
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_-_170744458
|
6.442
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr2_-_73869493
|
6.336
|
NM_003960
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative)
|
|
chr8_+_120079423
|
6.325
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr15_+_58430394
|
6.196
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr4_+_100485239
|
5.888
|
NM_000253
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_+_207262211
|
5.863
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr1_-_230849847
|
5.764
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr10_-_95360959
|
5.625
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr2_-_31637553
|
5.614
|
|
XDH
|
xanthine dehydrogenase
|
|
chr10_+_5238756
|
5.528
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr16_-_55866972
|
5.351
|
|
CES1
|
carboxylesterase 1
|
|
chr12_+_16500075
|
5.324
|
NM_145792
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr9_+_111624545
|
5.287
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chr16_-_55867016
|
5.204
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr17_-_17485744
|
5.065
|
NM_007169
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr4_+_74269971
|
5.014
|
NM_000477
|
ALB
|
albumin
|
|
chr5_-_147211235
|
4.869
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr3_-_190580464
|
4.662
|
NM_001146686
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr12_-_105322056
|
4.614
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr17_-_17485691
|
4.549
|
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_+_196946666
|
4.278
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr10_-_104597079
|
4.097
|
NM_000102
|
CYP17A1
|
cytochrome P450, family 17, subfamily A, polypeptide 1
|
|
chr4_-_185747183
|
4.094
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr3_+_186330711
|
4.082
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr3_-_170744529
|
4.048
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr11_-_18270167
|
3.974
|
NM_001199744
NM_001127380
NM_030754
|
SAA2-SAA4
SAA2
|
SAA2-SAA4 readthrough
serum amyloid A2
|
|
chr5_-_140012730
|
3.963
|
|
CD14
|
CD14 molecule
|
|
chr17_-_34329083
|
3.944
|
NM_032965
|
CCL15
|
chemokine (C-C motif) ligand 15
|
|
chr9_-_104198045
|
3.894
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr6_-_133035180
|
3.854
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr8_+_24298442
|
3.850
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr17_-_64225496
|
3.798
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr12_-_88423145
|
3.685
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr5_-_140013034
|
3.619
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr8_-_6735438
|
3.614
|
NM_005218
|
DEFB1
|
defensin, beta 1
|
|
chr6_+_31895201
|
3.573
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr11_+_69061599
|
3.519
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr7_+_129847699
|
3.424
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr4_+_71200670
|
3.337
|
NM_033122
|
CABS1
|
calcium-binding protein, spermatid-specific 1
|
|
chr1_+_241695433
|
3.332
|
NM_003679
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr4_+_69962191
|
3.313
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr22_+_24979717
|
3.303
|
NM_013430
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr14_+_24563369
|
3.223
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr1_-_207143944
|
3.220
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr1_+_154377847
|
3.169
|
|
IL6R
|
interleukin 6 receptor
|
|
chr9_-_19033183
|
3.168
|
NM_153707
|
FAM154A
|
family with sequence similarity 154, member A
|
|
chr19_-_8809054
|
3.144
|
NM_178525
|
ACTL9
|
actin-like 9
|
|
chr20_+_55099784
|
3.118
|
NM_001012971
|
FAM209A
FAM209B
|
family with sequence similarity 209, member A
family with sequence similarity 209, member B
|
|
chr2_-_169887832
|
3.085
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr6_+_31895401
|
3.000
|
|
C2
|
complement component 2
|
|
chr4_+_39408472
|
2.967
|
NM_175737
|
KLB
|
klotho beta
|
|
chr11_-_66675334
|
2.946
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr8_-_145115565
|
2.940
|
NM_017570
|
OPLAH
|
5-oxoprolinase (ATP-hydrolysing)
|
|
chr14_+_103388992
|
2.932
|
NM_030943
|
AMN
|
amnionless homolog (mouse)
|
|
chr12_-_4754275
|
2.815
|
NM_006422
|
AKAP3
|
A kinase (PRKA) anchor protein 3
|
|
chr1_+_196621007
|
2.783
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr19_-_4540035
|
2.745
|
NM_052972
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr14_-_23284617
|
2.725
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr22_+_25003610
|
2.620
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr10_+_64893003
|
2.586
|
NM_030759
|
NRBF2
|
nuclear receptor binding factor 2
|
|
chr6_-_24489738
|
2.586
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr6_+_131148544
|
2.583
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|
|
chr19_+_45417920
|
2.517
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr4_-_170924911
|
2.498
|
NM_001009554
|
MFAP3L
|
microfibrillar-associated protein 3-like
|
|
chr9_+_103947316
|
2.482
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr4_-_185747051
|
2.477
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr8_+_10383055
|
2.470
|
NM_001197020
NM_198464
|
PRSS55
|
protease, serine, 55
|
|
chr1_+_209878135
|
2.455
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr20_+_23471765
|
2.397
|
NM_005492
|
CST8
|
cystatin 8 (cystatin-related epididymal specific)
|
|
chr7_+_45933096
|
2.391
|
|
|
|
|
chr19_-_4540078
|
2.373
|
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr14_-_23284565
|
2.346
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr1_+_57320442
|
2.312
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr17_-_34328530
|
2.288
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr19_-_46088084
|
2.279
|
NM_001017989
NM_025136
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr1_+_110655061
|
2.213
|
NM_203412
|
UBL4B
|
ubiquitin-like 4B
|
|
chr1_-_162346579
|
2.179
|
NM_182581
|
C1orf111
|
chromosome 1 open reading frame 111
|
|
chr1_-_230850335
|
2.100
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr12_+_16064105
|
2.078
|
NM_015954
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr12_-_13256572
|
2.076
|
NM_001080554
NM_001080555
NM_001206842
|
GSG1
|
germ cell associated 1
|
|
chr9_-_91267004
|
2.075
|
NM_001100111
|
LOC286238
|
uncharacterized LOC286238
|
|
chr10_-_101190201
|
2.023
|
NM_002079
|
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1)
|
|
chr11_+_112047088
|
2.012
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr19_+_45418063
|
2.012
|
|
APOC1
|
apolipoprotein C-I
|
|
chr1_-_153123426
|
2.010
|
NM_001014291
|
SPRR2G
|
small proline-rich protein 2G
|
|
chr16_+_68057144
|
2.003
|
NM_017803
|
DUS2L
|
dihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae)
|
|
chr17_-_39324423
|
1.975
|
NM_033187
|
KRTAP4-3
|
keratin associated protein 4-3
|
|
chr3_+_148447966
|
1.952
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr15_-_83621350
|
1.914
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr15_+_91411884
|
1.895
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr7_+_128349111
|
1.888
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr12_+_16064232
|
1.874
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr11_+_112046207
|
1.859
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr20_+_42187634
|
1.859
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr12_+_57623355
|
1.830
|
NM_001166356
NM_005412
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr5_+_42548314
|
1.830
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr1_-_204121144
|
1.827
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chr12_+_16064282
|
1.820
|
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr8_+_40010986
|
1.809
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr5_+_42425093
|
1.798
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr17_-_41277409
|
1.795
|
NM_007297
NM_007299
NM_007294
NM_007300
|
BRCA1
|
breast cancer 1, early onset
|
|
chr17_+_685512
|
1.771
|
NM_018146
|
RNMTL1
|
RNA methyltransferase like 1
|
|
chr4_+_74702272
|
1.762
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr1_+_196788860
|
1.748
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr1_+_152850797
|
1.747
|
NM_030663
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein
|
|
chr19_-_46088021
|
1.741
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr1_+_151739130
|
1.739
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr4_-_100485173
|
1.728
|
NM_001134665
NM_001134666
|
RG9MTD2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chrY_+_3447081
|
1.728
|
NM_139214
|
TGIF2LY
|
TGFB-induced factor homeobox 2-like, Y-linked
|
|
chr17_-_34328607
|
1.705
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chr2_-_21228189
|
1.698
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr20_+_30028410
|
1.672
|
NM_153324
|
DEFB123
|
defensin, beta 123
|
|
chr13_-_60325073
|
1.667
|
|
|
|
|
chr9_-_107666046
|
1.660
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_+_192127591
|
1.660
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr14_+_21423627
|
1.659
|
NM_002934
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin)
|
|
chr1_+_112016588
|
1.640
|
NM_174896
|
C1orf162
|
chromosome 1 open reading frame 162
|
|
chr18_+_54814292
|
1.636
|
|
BOD1P
|
biorientation of chromosomes in cell division 1 pseudogene
|
|
chr3_-_195938256
|
1.628
|
NM_001039617
|
ZDHHC19
|
zinc finger, DHHC-type containing 19
|
|
chr16_-_68057098
|
1.623
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr4_+_118955499
|
1.612
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr17_+_51900238
|
1.609
|
NM_032559
|
KIF2B
|
kinesin family member 2B
|
|
chr2_+_3705785
|
1.600
|
NM_018436
|
ALLC
|
allantoicase
|
|
chr2_-_27362277
|
1.593
|
NM_178553
|
C2orf53
|
chromosome 2 open reading frame 53
|
|
chr1_-_204183036
|
1.584
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr9_+_105757592
|
1.582
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr4_-_23891608
|
1.581
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_-_132032156
|
1.570
|
NM_001145659
|
CTAGE9
|
CTAGE family, member 9
|
|
chr2_+_211421322
|
1.562
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr7_+_143880547
|
1.536
|
NM_198495
|
CTAGE4
CTAGE8
|
CTAGE family, member 4
CTAGE family, member 8
|
|
chr1_+_209859524
|
1.535
|
NM_001206741
NM_181755
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr2_-_234652623
|
1.510
|
NM_001001394
|
DNAJB3
|
DnaJ (Hsp40) homolog, subfamily B, member 3
|
|
chr6_+_150690027
|
1.508
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chr19_-_46088074
|
1.502
|
|
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia)
|
|
chr10_+_89267589
|
1.492
|
NM_001178118
|
MINPP1
|
multiple inositol-polyphosphate phosphatase 1
|
|
chr12_+_57623827
|
1.474
|
NM_001166358
NM_001166359
NM_001166357
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial)
|
|
chr1_+_186265404
|
1.465
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chr1_-_204120909
|
1.447
|
|
ETNK2
|
ethanolamine kinase 2
|
|
chr9_+_110045516
|
1.446
|
NM_002874
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr7_-_122342965
|
1.443
|
NM_198085
|
RNF148
|
ring finger protein 148
|
|
chr6_-_161087406
|
1.442
|
NM_005577
|
LPA
|
lipoprotein, Lp(a)
|
|
chr12_-_7261788
|
1.428
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr16_-_30457197
|
1.420
|
NM_012248
|
SEPHS2
|
selenophosphate synthetase 2
|
|
chr3_+_142342265
|
1.419
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr5_-_147162251
|
1.407
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr16_-_20911560
|
1.394
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chrY_-_15591799
|
1.391
|
|
UTY
|
ubiquitously transcribed tetratricopeptide repeat gene, Y-linked
|
|
chr16_-_68057120
|
1.381
|
|
DDX28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr20_-_1600589
|
1.375
|
NM_001083910
NM_001135844
NM_006065
|
SIRPB1
|
signal-regulatory protein beta 1
|
|
chr2_+_232457574
|
1.367
|
NM_152614
|
C2orf57
|
chromosome 2 open reading frame 57
|
|
chr1_-_203155910
|
1.366
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr9_-_104145800
|
1.354
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr17_-_39274587
|
1.348
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr11_-_68039327
|
1.347
|
|
C11orf24
|
chromosome 11 open reading frame 24
|
|
chr20_+_42187705
|
1.346
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr7_+_142636602
|
1.340
|
NM_178829
|
C7orf34
|
chromosome 7 open reading frame 34
|