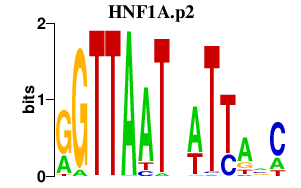
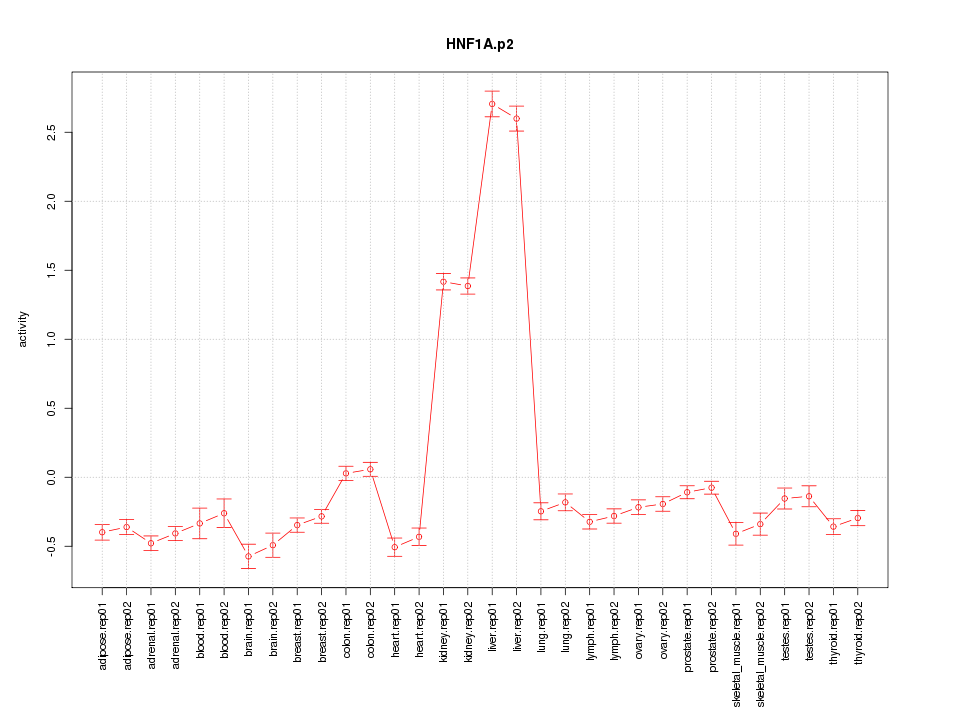
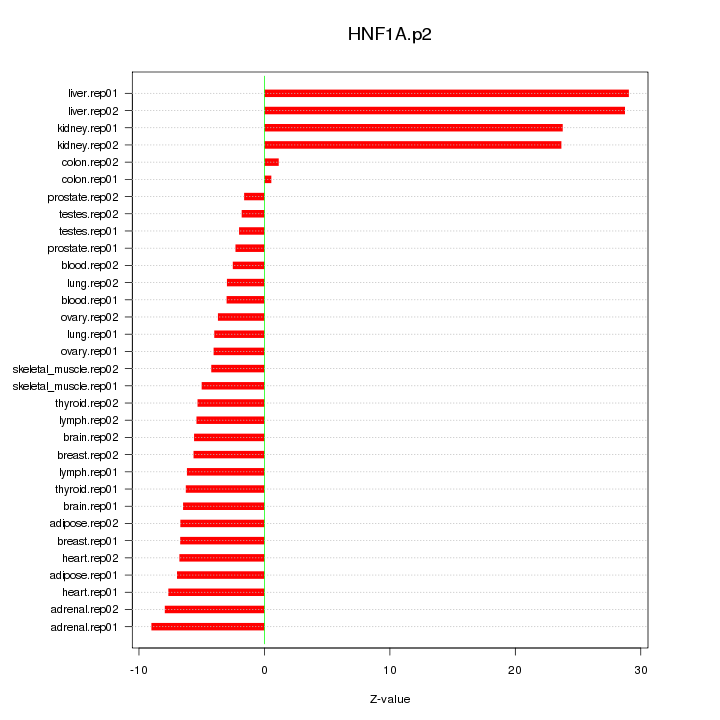
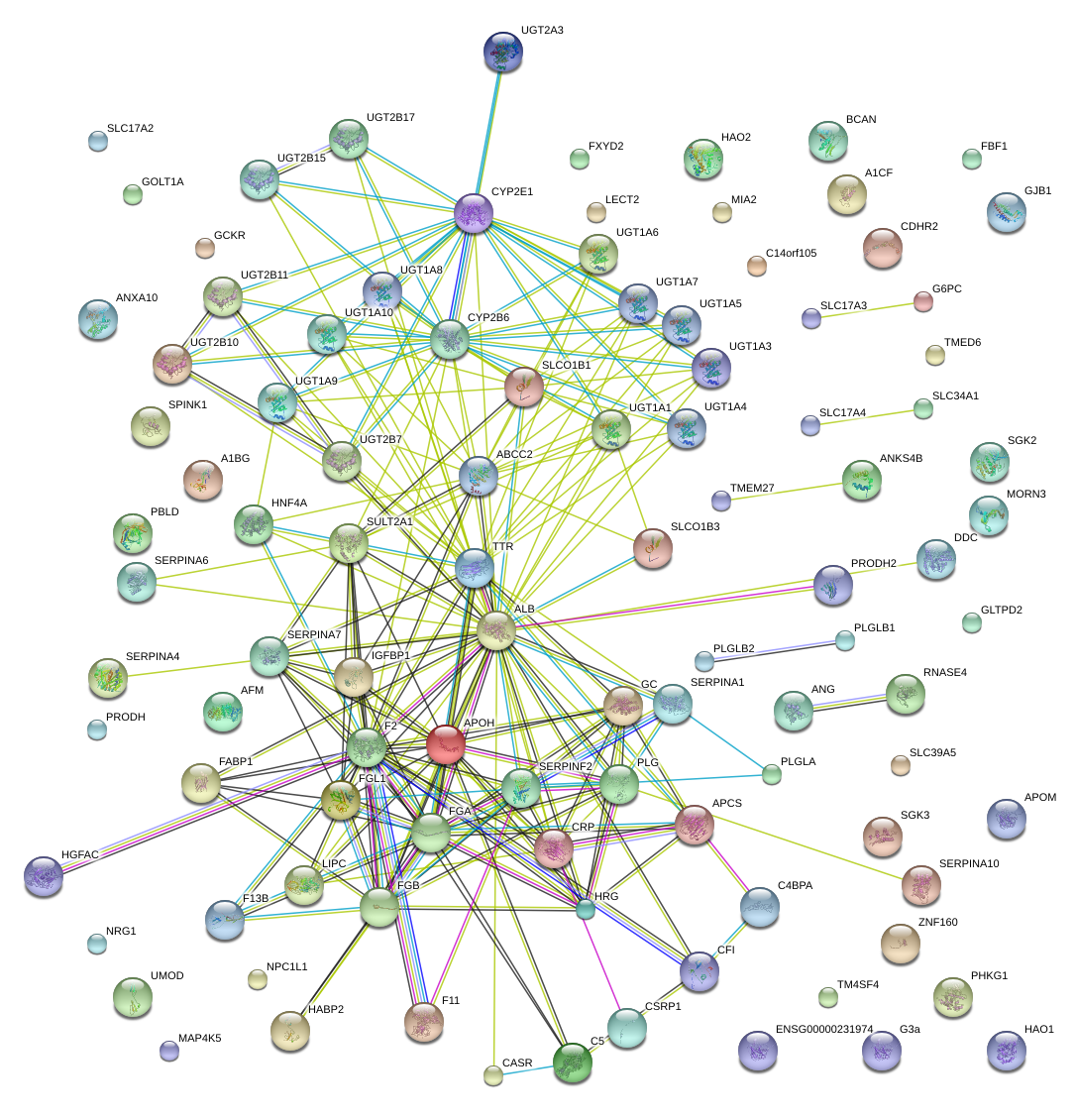
|
chr4_-_72649734
|
161.545
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr1_-_159684274
|
158.324
|
NM_000567
|
CRP
|
C-reactive protein, pentraxin-related
|
|
chr4_+_74347461
|
145.745
|
NM_001133
|
AFM
|
afamin
|
|
chr4_-_155511838
|
131.965
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr4_+_155484131
|
129.741
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr17_-_64225496
|
119.840
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr4_+_74269971
|
115.688
|
NM_000477
|
ALB
|
albumin
|
|
chr1_+_159557615
|
106.774
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr17_+_41052813
|
95.277
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr2_+_234668914
|
93.719
|
NM_000463
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1
|
|
chr5_-_147211141
|
91.777
|
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr2_+_234601511
|
89.928
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr19_-_36303766
|
89.070
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chrX_-_105282714
|
88.849
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr5_-_147211235
|
88.367
|
NM_003122
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1
|
|
chr8_-_17752847
|
88.288
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr14_-_94854910
|
86.867
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr6_+_161123224
|
86.262
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr14_-_94789641
|
82.717
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr14_-_94855120
|
81.964
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr20_+_43029865
|
81.254
|
NM_000457
NM_178849
NM_178850
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_-_36303724
|
78.575
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr1_+_119911395
|
77.735
|
NM_001005783
NM_016527
|
HAO2
|
hydroxyacid oxidase 2 (long chain)
|
|
chr2_+_234526290
|
76.621
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr14_+_95027760
|
76.456
|
NM_006215
|
SERPINA4
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4
|
|
chr12_+_21284127
|
73.205
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr1_-_197036350
|
71.328
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr14_-_57960432
|
70.934
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr20_-_7921068
|
70.369
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr2_+_234627423
|
69.508
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr14_-_94759594
|
66.478
|
NM_001100607
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr4_-_69817478
|
65.644
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr11_+_46740769
|
65.339
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr2_-_88427535
|
64.243
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr1_-_204183036
|
63.425
|
NM_198447
|
GOLT1A
|
golgi transport 1A
|
|
chr5_-_135290521
|
62.367
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr10_+_135340865
|
62.354
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr3_+_186383770
|
61.623
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr14_-_94759556
|
61.062
|
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr19_-_48389514
|
61.027
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr15_+_58724174
|
60.040
|
NM_000236
|
LIPC
|
lipase, hepatic
|
|
chr10_+_115312777
|
59.070
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr20_+_43030005
|
58.495
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr4_-_69434215
|
58.311
|
NM_001077
|
UGT2B15
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B15
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr14_-_94759360
|
57.949
|
NM_016186
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10
|
|
chr19_+_41509825
|
57.313
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr1_+_207277577
|
57.048
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr11_+_46740736
|
56.962
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr19_-_48389571
|
56.346
|
NM_003167
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr20_+_42984088
|
56.174
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr19_-_36304200
|
55.986
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr4_+_187187117
|
55.903
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr10_-_52645430
|
55.481
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr20_+_43029936
|
55.199
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr11_-_117695408
|
54.876
|
NM_001680
|
FXYD2
|
FXYD domain containing ion transport regulator 2
|
|
chrX_+_70435087
|
54.397
|
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr12_+_20963637
|
54.235
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr20_+_42984338
|
51.862
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr4_+_69962191
|
50.054
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr7_+_45927958
|
49.911
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr17_+_4692226
|
49.683
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr17_+_1646129
|
48.890
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chrX_+_70435048
|
48.629
|
NM_001097642
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr20_+_42187705
|
48.386
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr7_-_50633042
|
47.706
|
NM_001082971
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase)
|
|
chr6_+_31623688
|
47.200
|
|
APOM
|
apolipoprotein M
|
|
chr6_+_31623668
|
45.872
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr20_+_43029973
|
45.528
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr16_+_21245015
|
45.259
|
NM_145865
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr2_+_234637772
|
44.719
|
NM_019093
|
UGT1A3
|
UDP glucuronosyltransferase 1 family, polypeptide A3
|
|
chr9_-_123812535
|
44.573
|
NM_001735
|
C5
|
complement component 5
|
|
chr4_-_110723127
|
44.384
|
|
CFI
|
complement factor I
|
|
chr7_-_44580857
|
44.210
|
NM_001101648
NM_013389
|
NPC1L1
|
NPC1 (Niemann-Pick disease, type C1, gene)-like 1
|
|
chr10_+_99344421
|
43.213
|
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr14_+_39703119
|
42.377
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr2_+_88047603
|
42.170
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr10_+_101542460
|
42.105
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr19_+_11350282
|
41.450
|
NM_018687
|
C19orf80
|
chromosome 19 open reading frame 80
|
|
chr3_+_149192367
|
40.344
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr20_+_42187634
|
39.939
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr17_+_1646143
|
39.601
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chrX_-_15683153
|
38.694
|
NM_020665
|
TMEM27
|
transmembrane protein 27
|
|
chr18_+_29171729
|
38.455
|
NM_000371
|
TTR
|
transthyretin
|
|
chr3_+_149192479
|
37.816
|
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr11_+_46740759
|
37.719
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr19_-_58864820
|
36.944
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr14_+_21156922
|
36.708
|
NM_001097577
NM_194431
|
ANG
RNASE4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family, 4
|
|
chr2_-_87248942
|
36.641
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr10_-_70066594
|
36.529
|
|
PBLD
|
phenazine biosynthesis-like protein domain containing
|
|
chr6_-_25874439
|
36.084
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr2_+_234580504
|
36.020
|
NM_021027
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr10_-_52645384
|
34.214
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr4_-_110723334
|
34.152
|
NM_000204
|
CFI
|
complement factor I
|
|
chr6_-_25930838
|
33.452
|
NM_005835
|
SLC17A2
|
solute carrier family 17 (sodium phosphate), member 2
|
|
chr2_+_27719702
|
32.791
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr5_+_175976342
|
32.223
|
NM_017675
|
CDHR2
|
cadherin-related family member 2
|
|
chr4_+_187187342
|
32.134
|
|
F11
|
coagulation factor XI
|
|
chr4_+_169013687
|
31.741
|
NM_007193
|
ANXA10
|
annexin A10
|
|
chr6_+_25754926
|
31.733
|
NM_005495
|
SLC17A4
|
solute carrier family 17 (sodium phosphate), member 4
|
|
chr4_+_3443691
|
30.788
|
NM_001528
|
HGFAC
|
HGF activator
|
|
chr10_+_101542554
|
30.391
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr17_+_1646290
|
28.989
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr16_-_20364029
|
28.947
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr16_-_69385656
|
28.935
|
NM_144676
|
TMED6
|
transmembrane emp24 protein transport domain containing 6
|
|
chr12_+_56624500
|
28.680
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr10_+_99344079
|
28.546
|
NM_001134670
NM_138413
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr2_+_234621637
|
28.460
|
NM_019078
|
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr5_-_138718941
|
28.426
|
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr16_-_87970107
|
27.383
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr17_+_73642645
|
27.252
|
NM_001162997
|
C17orf110
|
chromosome 17 open reading frame 110
|
|
chr4_+_3443659
|
26.835
|
|
HGFAC
|
HGF activator
|
|
chr1_+_18807423
|
25.954
|
NM_152375
|
KLHDC7A
|
kelch domain containing 7A
|
|
chr15_-_90349819
|
25.848
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr8_+_76320106
|
25.314
|
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr8_-_124749573
|
24.974
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr15_+_48498497
|
24.097
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr11_+_64323089
|
23.089
|
NM_018484
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11
|
|
chr19_-_33360641
|
22.835
|
NM_001126335
NM_001243036
NM_014270
|
SLC7A9
|
solute carrier family 7 (glycoprotein-associated amino acid transporter light chain, bo,+ system), member 9
|
|
chr6_-_116381765
|
21.689
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr11_+_114167168
|
21.680
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr15_+_58430394
|
21.531
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr1_+_207262211
|
21.293
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr16_-_48269087
|
20.966
|
NM_032583
NM_145186
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr12_-_113841654
|
20.948
|
NM_006843
|
SDS
|
serine dehydratase
|
|
chr6_-_51952368
|
20.386
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr9_-_116861336
|
19.552
|
NM_138424
|
KIF12
|
kinesin family member 12
|
|
chr6_-_24383519
|
19.375
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr10_+_5136613
|
19.039
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr1_-_20141593
|
18.570
|
NM_019062
|
RNF186
|
ring finger protein 186
|
|
chr10_+_5136589
|
18.399
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr9_-_116861506
|
18.229
|
|
KIF12
|
kinesin family member 12
|
|
chr17_+_73553883
|
17.894
|
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila)
|
|
chr4_-_69536314
|
17.513
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr1_+_162351519
|
17.454
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr5_-_138719018
|
16.987
|
NM_005847
NM_152685
|
SLC23A1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr10_+_5136556
|
16.948
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr6_+_131894343
|
16.636
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr3_+_190105660
|
16.341
|
NM_006580
|
CLDN16
|
claudin 16
|
|
chr6_-_52628272
|
15.668
|
NM_000846
|
GSTA2
|
glutathione S-transferase alpha 2
|
|
chr14_+_68168602
|
15.312
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr4_-_70080448
|
14.990
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr10_-_129691202
|
14.825
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr11_+_561449
|
14.771
|
NM_001143994
NM_003475
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr3_+_119499330
|
14.467
|
NM_003889
NM_033013
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr4_+_74270832
|
14.268
|
|
ALB
|
albumin
|
|
chr11_-_26743545
|
13.919
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr6_+_54173202
|
13.809
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr4_-_100273831
|
13.683
|
NM_000669
|
ADH1C
|
alcohol dehydrogenase 1C (class I), gamma polypeptide
|
|
chr12_+_10331556
|
13.363
|
NM_153022
|
C12orf59
|
chromosome 12 open reading frame 59
|
|
chr18_-_48346297
|
13.245
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr1_-_47407094
|
13.215
|
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11
|
|
chr4_-_100212085
|
13.192
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr6_+_167704802
|
13.169
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr11_+_114167197
|
12.814
|
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr2_-_99917638
|
12.734
|
NM_174898
|
LYG1
|
lysozyme G-like 1
|
|
chr19_+_45417576
|
12.376
|
|
APOC1
|
apolipoprotein C-I
|
|
chr11_-_118900078
|
12.255
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr21_+_39628869
|
12.174
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr1_-_230849936
|
12.066
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr12_+_121416294
|
11.989
|
|
HNF1A
|
HNF1 homeobox A
|
|
chr22_+_32439018
|
11.910
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr14_+_88490893
|
11.485
|
|
LOC283587
|
uncharacterized LOC283587
|
|
chr16_+_20775311
|
11.469
|
NM_005622
NM_202000
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3
|
|
chr1_+_47603083
|
11.347
|
NM_001010969
|
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22
|
|
chr20_+_56136136
|
11.319
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr1_+_94057518
|
11.318
|
|
LOC100129046
|
uncharacterized LOC100129046
|
|
chr2_+_182850550
|
11.208
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr8_-_6735438
|
11.131
|
NM_005218
|
DEFB1
|
defensin, beta 1
|
|
chr3_-_157221127
|
10.983
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr1_-_47407142
|
10.898
|
NM_000778
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11
|
|
chr1_+_94057581
|
10.662
|
|
LOC100129046
|
uncharacterized LOC100129046
|
|
chr19_-_39443326
|
10.617
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr4_+_71494460
|
10.579
|
NM_031889
|
ENAM
|
enamelin
|
|
chr15_+_59903980
|
10.314
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr21_+_39628663
|
10.301
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr11_-_62783281
|
10.285
|
NM_001184732
NM_001184733
NM_001184736
NM_004254
|
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8
|
|
chr10_+_96698409
|
10.278
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr22_+_32439250
|
10.225
|
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr6_+_64231626
|
10.113
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr4_-_987163
|
9.759
|
NM_022042
NM_134425
NM_213613
|
SLC26A1
|
solute carrier family 26 (sulfate transporter), member 1
|
|
chr7_-_107443631
|
9.422
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr12_-_102874158
|
9.280
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr16_-_55909208
|
9.054
|
NM_001143685
NM_145024
|
CES5A
|
carboxylesterase 5A
|
|
chr13_-_101185944
|
8.919
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr3_-_169587598
|
8.816
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr4_-_40516567
|
8.562
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr7_-_122839999
|
8.505
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr2_-_162931051
|
8.438
|
NM_001935
|
DPP4
|
dipeptidyl-peptidase 4
|
|
chr1_+_54359859
|
8.379
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr5_+_176513819
|
8.350
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr20_-_22564893
|
8.338
|
|
FOXA2
|
forkhead box A2
|
|
chr16_-_66952729
|
8.173
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr17_-_202617
|
7.876
|
NM_001190411
NM_001190412
NM_001190413
NM_006987
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr12_-_8693391
|
7.690
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr7_+_141695678
|
7.610
|
NM_004668
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase)
|
|
chr4_+_74301932
|
7.586
|
NM_001134
|
AFP
|
alpha-fetoprotein
|
|
chr17_-_202557
|
7.464
|
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chrX_-_131547536
|
7.367
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr17_+_68071386
|
7.211
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr1_+_199996729
|
7.195
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|