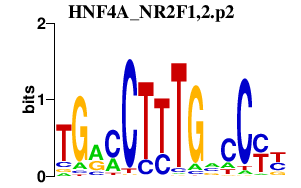
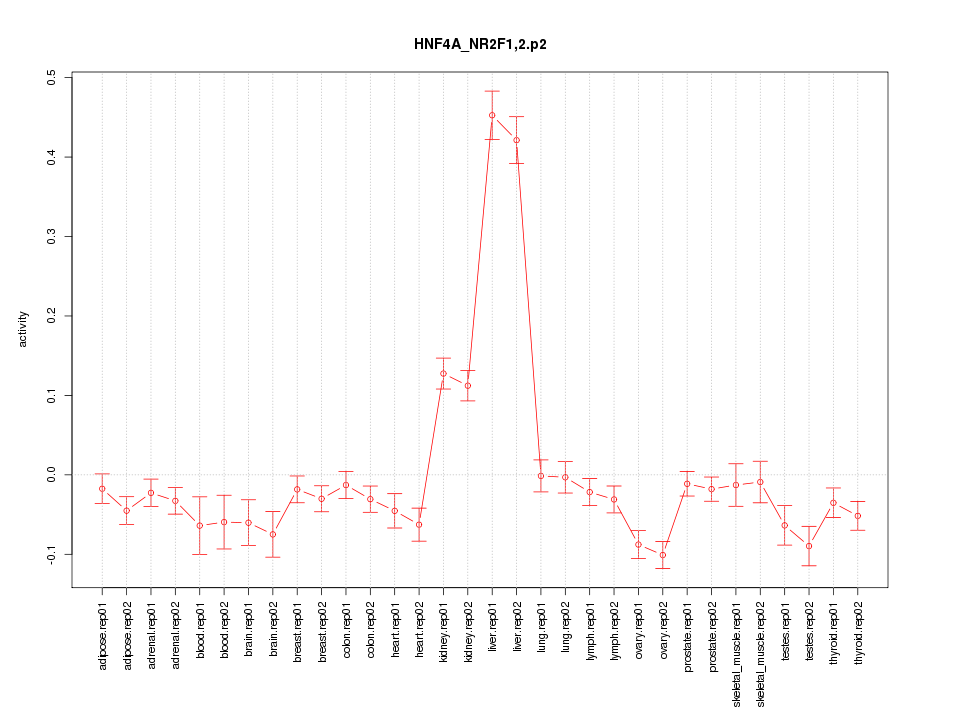
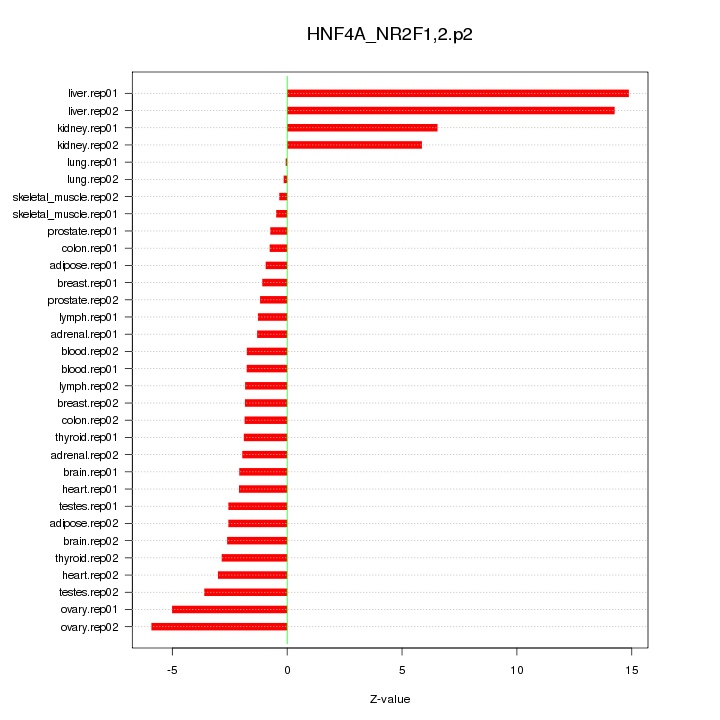
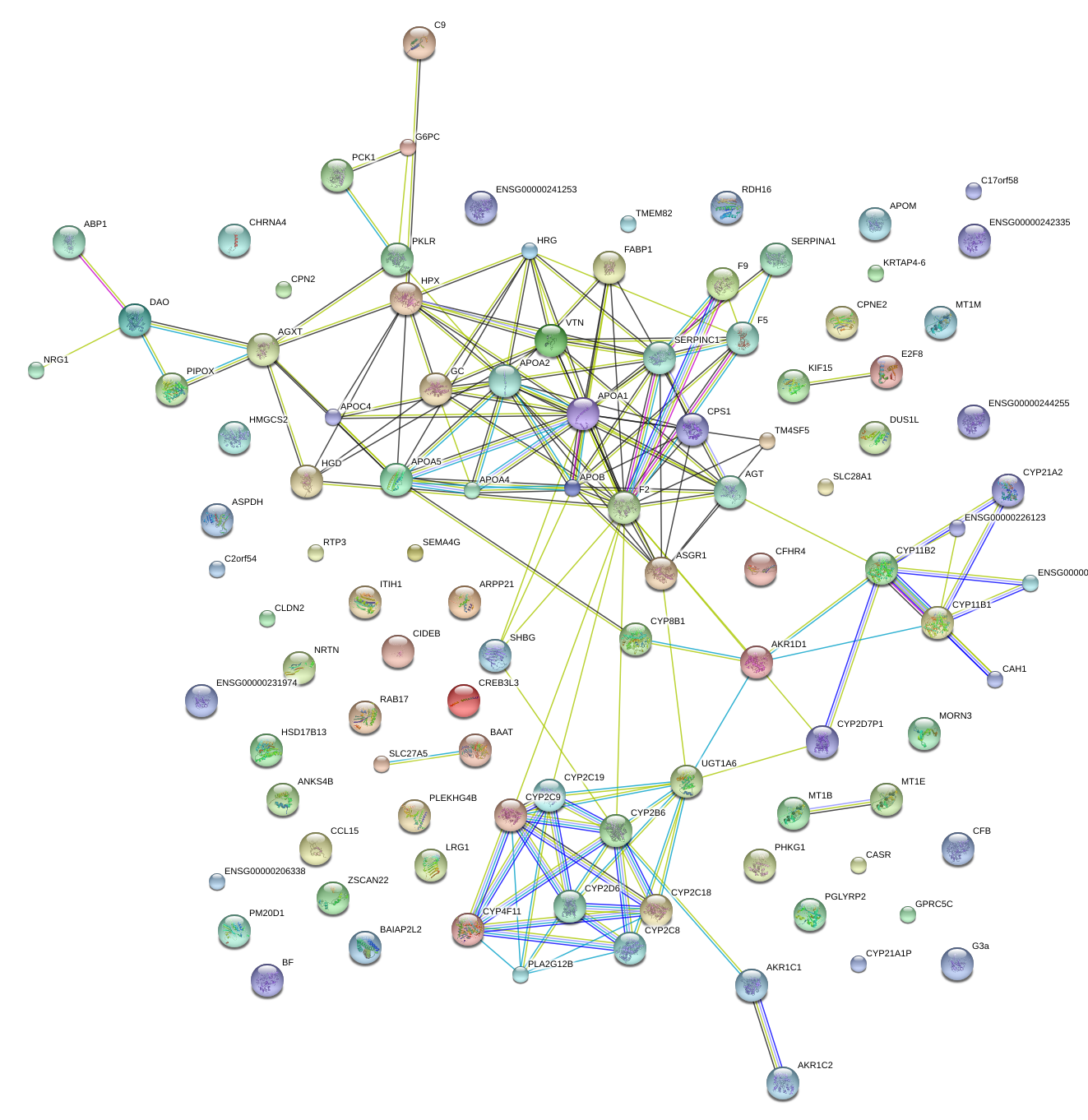
|
chr19_+_4153593
|
26.167
|
NM_032607
|
CREB3L3
|
cAMP responsive element binding protein 3-like 3
|
|
chr10_+_96698409
|
23.530
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr3_+_186383770
|
22.171
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr17_+_27370155
|
22.155
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr1_+_16068916
|
22.083
|
NM_001013641
|
TMEM82
|
transmembrane protein 82
|
|
chr3_-_42917191
|
21.035
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chrX_+_138612894
|
20.264
|
NM_000133
|
F9
|
coagulation factor IX
|
|
chr12_-_57351381
|
20.012
|
NM_003708
|
RDH16
|
retinol dehydrogenase 16 (all-trans)
|
|
chr1_-_173886464
|
19.728
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr3_-_194072004
|
19.203
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr3_+_52812438
|
18.730
|
NM_001166434
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1
|
|
chr1_-_173886401
|
18.457
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr16_+_56685810
|
17.782
|
NM_005947
|
MT1B
|
metallothionein 1B
|
|
chr4_+_22694547
|
17.119
|
NM_001128432
NM_020973
|
GBA3
|
glucosidase, beta, acid 3 (cytosolic)
|
|
chr14_-_94855120
|
17.079
|
NM_000295
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr10_+_96522457
|
16.296
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr15_+_85427891
|
16.280
|
NM_004213
NM_201651
|
SLC28A1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1
|
|
chr6_+_131148544
|
16.177
|
NM_001195597
|
LOC100507203
|
uncharacterized LOC100507203
|
|
chr17_-_34329083
|
15.665
|
NM_032965
|
CCL15
|
chemokine (C-C motif) ligand 15
|
|
chr11_-_6462129
|
15.065
|
NM_000613
|
HPX
|
hemopexin
|
|
chr1_-_161193190
|
14.975
|
|
APOA2
|
apolipoprotein A-II
|
|
chr19_-_15590203
|
14.252
|
NM_052890
|
PGLYRP2
|
peptidoglycan recognition protein 2
|
|
chr3_+_46539484
|
14.143
|
NM_031440
|
RTP3
|
receptor (chemosensory) transporter protein 3
|
|
chr11_-_116693907
|
13.664
|
NM_000482
|
APOA4
|
apolipoprotein A-IV
|
|
chr6_+_31913763
|
13.319
|
|
CFB
|
complement factor B
|
|
chrX_+_106143393
|
13.206
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr10_+_102732285
|
13.194
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr17_-_26697224
|
12.794
|
|
VTN
|
vitronectin
|
|
chr11_-_116663106
|
12.783
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr2_-_238499317
|
12.779
|
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr6_+_31913852
|
12.424
|
|
CFB
|
complement factor B
|
|
chr10_-_96829074
|
12.221
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr1_+_196857143
|
12.084
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr11_+_46740736
|
11.906
|
NM_000506
|
F2
|
coagulation factor II (thrombin)
|
|
chr11_+_46740769
|
11.854
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr17_+_4675175
|
11.846
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr17_+_27369917
|
11.772
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr17_+_41052813
|
11.553
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr1_-_161193375
|
11.508
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr2_-_238499713
|
11.436
|
NM_022449
|
RAB17
|
RAB17, member RAS oncogene family
|
|
chr1_-_230849847
|
11.231
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_-_230849863
|
11.160
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr5_-_39364562
|
11.127
|
NM_001737
|
C9
|
complement component 9
|
|
chrX_+_106171328
|
11.086
|
|
CLDN2
|
claudin 2
|
|
chr17_+_72439187
|
11.080
|
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr2_-_241831385
|
11.050
|
NM_024861
|
C2orf54
|
chromosome 2 open reading frame 54
|
|
chr3_-_42917632
|
11.048
|
NM_004391
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr10_+_96443250
|
11.029
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr19_-_4540078
|
10.838
|
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr2_-_21266893
|
10.833
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr12_+_109273856
|
10.736
|
NM_001917
|
DAO
|
D-amino-acid oxidase
|
|
chr1_-_155271186
|
10.622
|
NM_000298
|
PKLR
|
pyruvate kinase, liver and RBC
|
|
chr22_-_42526792
|
10.587
|
|
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6
|
|
chr20_-_61992516
|
10.564
|
NM_000744
|
CHRNA4
|
cholinergic receptor, nicotinic, alpha 4
|
|
chr20_+_56136136
|
10.344
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr6_+_31623668
|
10.336
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr22_-_42526877
|
10.335
|
NM_000106
NM_001025161
|
CYP2D6
|
cytochrome P450, family 2, subfamily D, polypeptide 6
|
|
chr4_-_72671236
|
10.269
|
NM_001204306
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr19_+_45445494
|
10.173
|
NM_001646
|
APOC4-APOC2
APOC4
|
APOC4-APOC2 readthrough
apolipoprotein C-IV
|
|
chr1_-_120311466
|
10.168
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr17_+_7533452
|
9.951
|
NM_001040
NM_001146279
NM_001146280
NM_001146281
|
SHBG
|
sex hormone-binding globulin
|
|
chr16_+_21245015
|
9.876
|
NM_145865
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr4_-_88243994
|
9.841
|
NM_001136230
NM_178135
|
HSD17B13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr6_+_31623688
|
9.837
|
|
APOM
|
apolipoprotein M
|
|
chr1_-_169555710
|
9.827
|
NM_000130
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr1_-_169555624
|
9.826
|
|
F5
|
coagulation factor V (proaccelerin, labile factor)
|
|
chr19_-_59023323
|
9.810
|
NM_012254
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5
|
|
chr2_-_21225640
|
9.698
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr17_+_7531275
|
9.646
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr19_+_5823817
|
9.616
|
NM_004558
|
NRTN
|
neurturin
|
|
chr2_-_88427535
|
9.518
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr7_+_137761177
|
9.466
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr16_+_56666533
|
9.431
|
NM_176870
|
MT1M
|
metallothionein 1M
|
|
chr10_-_74714479
|
9.293
|
NM_032562
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr11_-_116708334
|
9.246
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr17_-_7082654
|
9.140
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr14_-_24777064
|
9.119
|
|
|
|
|
chr19_+_41497182
|
9.091
|
NM_000767
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr19_-_51017124
|
9.085
|
NM_001114598
|
ASPDH
|
aspartate dehydrogenase domain containing
|
|
chr14_-_24777160
|
8.983
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr14_-_94854910
|
8.878
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr19_-_16045669
|
8.617
|
NM_001128932
NM_021187
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11
|
|
chr3_-_120400957
|
8.579
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr9_-_104147261
|
8.548
|
NM_001701
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr1_-_230850335
|
8.536
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr14_-_24777153
|
8.457
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr2_+_211421322
|
8.438
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr6_+_31982538
|
8.414
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr22_+_24891114
|
8.412
|
NM_016327
|
UPB1
|
ureidopropionase, beta
|
|
chr8_-_17743062
|
8.409
|
|
FGL1
|
fibrinogen-like 1
|
|
chr19_+_41594367
|
8.375
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr3_+_186435097
|
8.347
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr12_-_21757752
|
8.218
|
NM_021957
|
GYS2
|
glycogen synthase 2 (liver)
|
|
chr4_+_69962191
|
8.200
|
NM_001074
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7
|
|
chr14_-_24777184
|
8.160
|
|
CIDEB
|
cell death-inducing DFFA-like effector b
|
|
chr6_+_31913708
|
8.053
|
NM_001710
|
CFB
|
complement factor B
|
|
chr6_+_31913920
|
8.020
|
|
CFB
|
complement factor B
|
|
chr13_+_113760101
|
7.957
|
NM_000131
NM_019616
|
F7
|
coagulation factor VII (serum prothrombin conversion accelerator)
|
|
chr2_+_27719702
|
7.942
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr1_-_177939049
|
7.941
|
NM_033127
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr14_-_65409445
|
7.861
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr19_-_39303520
|
7.812
|
|
LGALS4
|
lectin, galactoside-binding, soluble, 4
|
|
chr17_+_1646290
|
7.804
|
NM_001165920
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr19_-_7968426
|
7.764
|
|
|
|
|
chr10_+_74653338
|
7.764
|
NM_152635
|
OIT3
|
oncoprotein induced transcript 3
|
|
chr12_-_122296619
|
7.751
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr10_-_129691202
|
7.714
|
NM_152311
|
CLRN3
|
clarin 3
|
|
chr22_+_25003610
|
7.645
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr2_+_211458078
|
7.577
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr2_+_234601511
|
7.553
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr17_+_19437140
|
7.522
|
NM_018242
|
SLC47A1
|
solute carrier family 47, member 1
|
|
chr1_-_151345156
|
7.515
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr12_-_322612
|
7.501
|
NM_003044
NM_001122847
|
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12
|
|
chr10_-_74714563
|
7.420
|
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr11_+_46740759
|
7.409
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr17_-_26697293
|
7.387
|
|
VTN
|
vitronectin
|
|
chr4_+_100495972
|
7.342
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr2_+_10091791
|
7.312
|
NM_198182
|
GRHL1
|
grainyhead-like 1 (Drosophila)
|
|
chr19_+_41497284
|
7.233
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr11_+_63137260
|
7.228
|
NM_080866
|
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9
|
|
chr10_-_129691047
|
7.141
|
|
|
|
|
chr1_+_196946666
|
7.137
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chrX_+_106163605
|
7.114
|
NM_020384
|
CLDN2
|
claudin 2
|
|
chr6_+_43266008
|
7.111
|
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7
|
|
chr13_+_113777112
|
7.040
|
NM_000504
|
F10
|
coagulation factor X
|
|
chr12_-_56756526
|
6.970
|
NM_001638
|
APOF
|
apolipoprotein F
|
|
chr1_+_171060017
|
6.861
|
NM_001002294
NM_006894
|
FMO3
|
flavin containing monooxygenase 3
|
|
chr12_+_100897137
|
6.835
|
NM_001206992
NM_001206993
|
NR1H4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr17_-_26697310
|
6.831
|
|
VTN
|
vitronectin
|
|
chr17_+_7533403
|
6.783
|
|
SHBG
|
sex hormone-binding globulin
|
|
chrX_-_105282714
|
6.780
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr3_+_186330711
|
6.737
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr20_-_7921068
|
6.730
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr11_+_2924504
|
6.724
|
|
SLC22A18
|
solute carrier family 22, member 18
|
|
chr12_+_57828457
|
6.674
|
NM_005538
|
INHBC
|
inhibin, beta C
|
|
chr1_-_11107150
|
6.650
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr6_+_31895401
|
6.614
|
|
C2
|
complement component 2
|
|
chr17_-_36105005
|
6.581
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr16_-_16317290
|
6.540
|
NM_001079528
NM_001171
|
ABCC6
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6
|
|
chr17_-_26697358
|
6.506
|
NM_000638
|
VTN
|
vitronectin
|
|
chr17_-_7018049
|
6.474
|
NM_001181
NM_001201352
NM_080912
NM_080913
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr19_-_36304200
|
6.462
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr10_+_115310589
|
6.429
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr2_+_219283809
|
6.376
|
NM_007127
|
VIL1
|
villin 1
|
|
chr17_+_1646143
|
6.367
|
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr3_+_108855560
|
6.364
|
|
FLJ22763
|
uncharacterized LOC401081
|
|
chr1_-_155270738
|
6.330
|
NM_181871
|
PKLR
|
pyruvate kinase, liver and RBC
|
|
chr17_-_34328530
|
6.321
|
|
CCL14-CCL15
CCL15
|
CCL14-CCL15 readthrough
chemokine (C-C motif) ligand 15
|
|
chr19_-_4540035
|
6.293
|
NM_052972
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr17_+_1646129
|
6.284
|
NM_000934
NM_001165921
|
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
|
|
chr12_+_16500528
|
6.258
|
NM_020300
NM_145791
|
MGST1
|
microsomal glutathione S-transferase 1
|
|
chr1_-_230849936
|
6.160
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr22_-_30901597
|
6.148
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr4_-_175443791
|
6.115
|
NM_000860
NM_001145816
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr10_+_101542554
|
6.008
|
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr7_-_15601574
|
5.967
|
NM_001004320
|
AGMO
|
alkylglycerol monooxygenase
|
|
chr11_-_62996998
|
5.962
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr3_-_50340979
|
5.941
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr2_+_219646448
|
5.888
|
NM_000784
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr5_-_42811959
|
5.841
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr7_-_100240334
|
5.816
|
|
TFR2
|
transferrin receptor 2
|
|
chr1_-_24127157
|
5.745
|
NM_001008216
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr11_-_61348290
|
5.710
|
NM_001252065
NM_004200
|
SYT7
|
synaptotagmin VII
|
|
chr1_+_196788860
|
5.695
|
NM_002113
|
CFHR1
|
complement factor H-related 1
|
|
chr11_+_74870824
|
5.690
|
NM_001145211
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr7_-_99277518
|
5.654
|
NM_000777
NM_001190484
|
CYP3A5
|
cytochrome P450, family 3, subfamily A, polypeptide 5
|
|
chr1_-_24126891
|
5.642
|
NM_000403
|
GALE
|
UDP-galactose-4-epimerase
|
|
chr10_+_101542460
|
5.637
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr12_+_113860146
|
5.613
|
NM_138432
|
SDSL
|
serine dehydratase-like
|
|
chr1_-_177939347
|
5.612
|
|
SEC16B
|
SEC16 homolog B (S. cerevisiae)
|
|
chr17_+_4692226
|
5.553
|
NM_001014985
|
GLTPD2
|
glycolipid transfer protein domain containing 2
|
|
chr17_-_7017971
|
5.492
|
|
ASGR2
|
asialoglycoprotein receptor 2
|
|
chr12_-_7125727
|
5.473
|
NM_005768
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3
|
|
chr16_-_87970107
|
5.431
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr6_+_31949800
|
5.420
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr2_+_158958381
|
5.402
|
NM_173355
|
UPP2
|
uridine phosphorylase 2
|
|
chr1_-_15911461
|
5.383
|
NM_024758
|
AGMAT
|
agmatine ureohydrolase (agmatinase)
|
|
chr4_-_175443601
|
5.370
|
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD)
|
|
chr9_+_74764266
|
5.368
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr5_-_41213513
|
5.354
|
NM_000065
|
C6
|
complement component 6
|
|
chr2_+_120125141
|
5.353
|
NM_001079863
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein)
|
|
chr3_-_58522873
|
5.347
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr10_-_50970321
|
5.331
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr1_+_145727712
|
5.321
|
|
PDZK1
|
PDZ domain containing 1
|
|
chr17_-_36104874
|
5.313
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr6_+_30130982
|
5.288
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr19_+_6464259
|
5.284
|
NM_139161
NM_174881
|
CRB3
|
crumbs homolog 3 (Drosophila)
|
|
chr19_+_45116921
|
5.275
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr1_+_145727665
|
5.264
|
NM_001201325
NM_001201326
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr12_+_56075418
|
5.242
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr17_-_34308455
|
5.235
|
NM_004590
|
CCL16
|
chemokine (C-C motif) ligand 16
|
|
chr2_-_165477818
|
5.211
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr2_-_31637598
|
5.173
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chrX_+_38211735
|
5.169
|
NM_000531
|
OTC
|
ornithine carbamoyltransferase
|
|
chr12_+_56075329
|
5.129
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr2_-_21266809
|
5.107
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr2_-_169887832
|
5.094
|
NM_003742
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr10_+_135160843
|
5.081
|
NM_001145201
NM_145202
|
PRAP1
|
proline-rich acidic protein 1
|
|
chr17_-_34328607
|
5.072
|
|
CCL14-CCL15
|
CCL14-CCL15 readthrough
|
|
chrX_-_128977291
|
5.041
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|