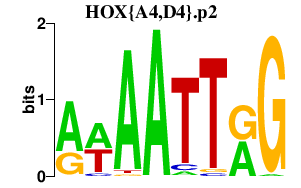
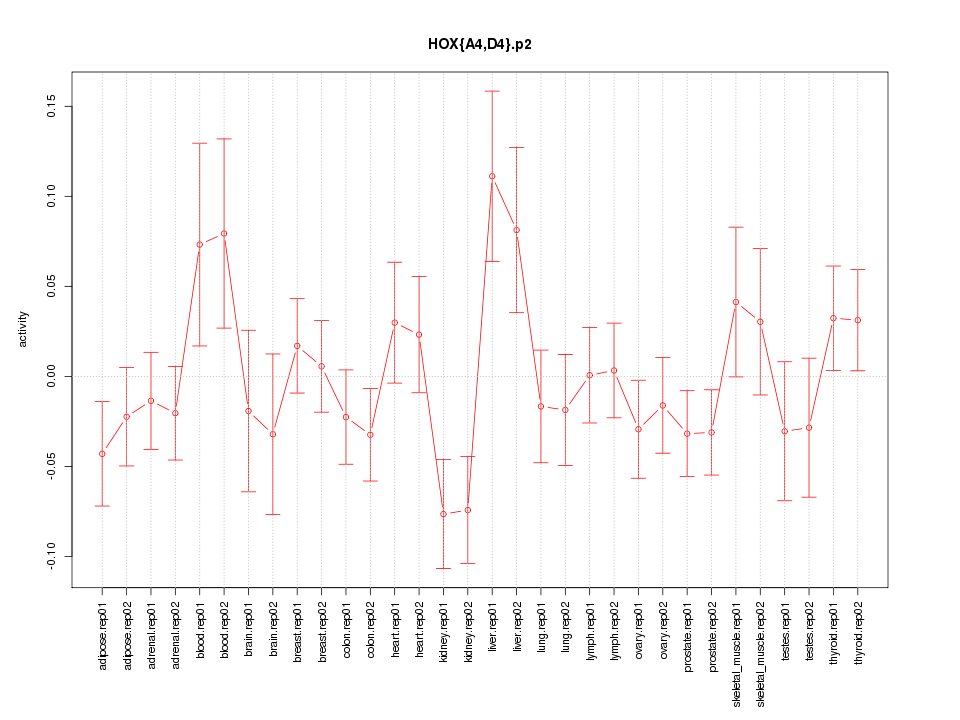
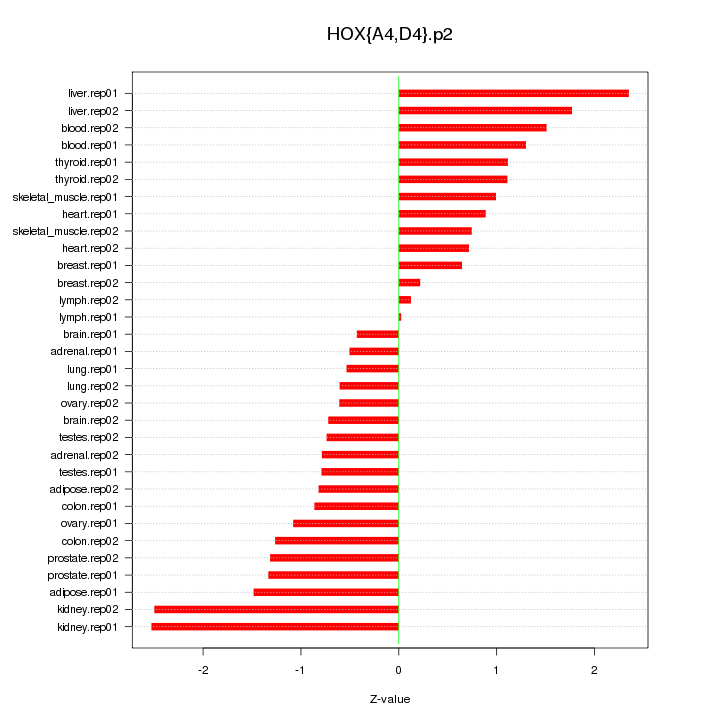
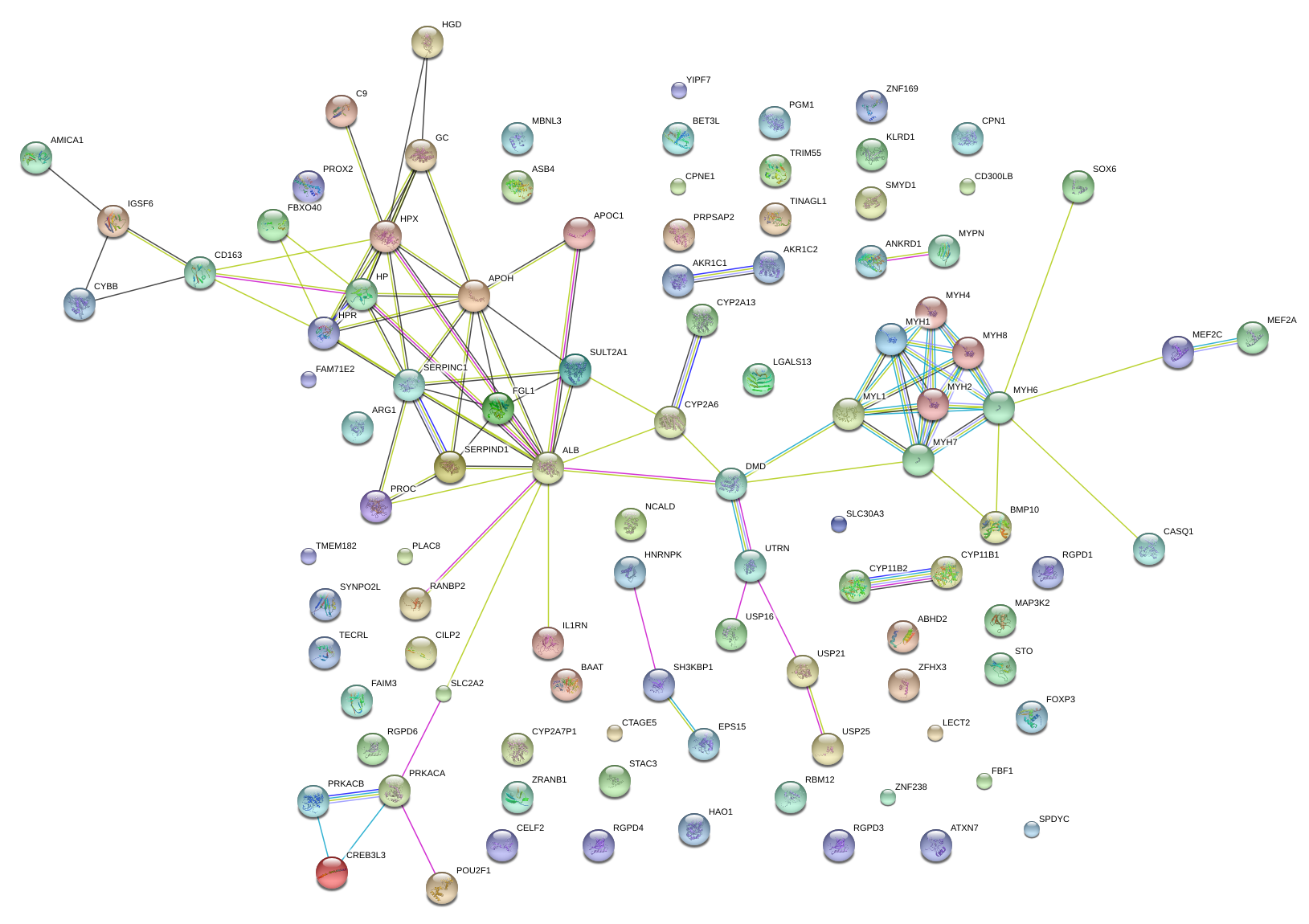
|
chr4_+_74269971
|
6.714
|
NM_000477
|
ALB
|
albumin
|
|
chr8_-_17743062
|
4.404
|
|
FGL1
|
fibrinogen-like 1
|
|
chr10_+_11047339
|
4.360
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr11_-_6462129
|
3.682
|
NM_000613
|
HPX
|
hemopexin
|
|
chr4_-_72649734
|
3.671
|
NM_000583
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr10_+_11047253
|
3.323
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr10_-_92681025
|
3.269
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_-_104145800
|
2.917
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr4_-_65275000
|
2.908
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr10_+_11047274
|
2.813
|
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr4_-_44653657
|
2.785
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chrX_-_131573638
|
2.719
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr17_-_64225496
|
2.607
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr17_-_10421858
|
2.539
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chrX_-_19688991
|
2.497
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_167298280
|
2.382
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr3_+_121312169
|
2.371
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr11_-_16424391
|
2.341
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr22_+_21128378
|
2.327
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr17_-_10452939
|
2.315
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr5_-_39364562
|
2.298
|
NM_001737
|
C9
|
complement component 9
|
|
chr10_+_126630691
|
2.181
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr2_+_128175989
|
2.147
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr8_+_67039130
|
2.141
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr5_-_135290521
|
2.132
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr19_+_4153593
|
2.094
|
NM_032607
|
CREB3L3
|
cAMP responsive element binding protein 3-like 3
|
|
chrX_+_37639306
|
2.061
|
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr1_-_207095198
|
2.045
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr1_+_167298382
|
2.023
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr4_+_74270832
|
1.975
|
|
ALB
|
albumin
|
|
chr19_-_48389514
|
1.944
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr16_-_73092533
|
1.895
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr12_+_10460416
|
1.866
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr14_-_23904849
|
1.861
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr8_-_17752847
|
1.837
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr9_-_86584079
|
1.814
|
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K
|
|
chr2_+_103378489
|
1.807
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr12_-_57644905
|
1.774
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr19_-_48389571
|
1.745
|
NM_003167
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chrX_+_37639246
|
1.734
|
NM_000397
|
CYBB
|
cytochrome b-245, beta polypeptide
|
|
chr20_-_7921068
|
1.678
|
NM_017545
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1
|
|
chr3_+_63884074
|
1.669
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr3_-_170744767
|
1.664
|
NM_000340
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr2_+_87144737
|
1.663
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr16_-_53405040
|
1.649
|
NM_001207030
|
LOC643802
|
u3 small nucleolar ribonucleoprotein protein MPP10-like
|
|
chr3_-_120400957
|
1.620
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr2_+_113875469
|
1.616
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr14_+_39734475
|
1.606
|
NM_203354
|
CTAGE5
|
CTAGE family, member 5
|
|
chr10_+_69869249
|
1.597
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr19_+_45116921
|
1.584
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr19_-_41356193
|
1.570
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr10_-_92680784
|
1.557
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr17_-_72527592
|
1.546
|
NM_174892
|
CD300LB
|
CD300 molecule-like family member b
|
|
chr1_+_160160284
|
1.530
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr2_-_211179765
|
1.526
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr5_-_88119604
|
1.491
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr17_+_18768781
|
1.484
|
NM_001243940
|
PRPSAP2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2
|
|
chrX_-_49121171
|
1.474
|
NM_001114377
NM_014009
|
FOXP3
|
forkhead box P3
|
|
chr2_-_69098502
|
1.469
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr1_+_244216506
|
1.460
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr11_+_64937624
|
1.393
|
NM_001008778
|
SPDYC
|
speedy homolog C (Xenopus laevis)
|
|
chr12_-_7656285
|
1.389
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr15_+_89739234
|
1.375
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr20_-_34243118
|
1.355
|
|
RBM12
|
RNA binding motif protein 12
|
|
chr15_+_100173182
|
1.350
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr11_-_118084073
|
1.339
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr6_-_116866772
|
1.333
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr10_-_75415748
|
1.320
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr16_-_21663908
|
1.316
|
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr2_+_88367298
|
1.315
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr19_+_45417920
|
1.305
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr16_+_72097124
|
1.303
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr1_+_84630575
|
1.301
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_-_51887740
|
1.301
|
NM_001159969
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr2_-_128100673
|
1.297
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chrX_-_33357725
|
1.281
|
NM_000109
|
DMD
|
dystrophin
|
|
chr5_+_176562084
|
1.280
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr10_-_101841593
|
1.270
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr7_+_95115212
|
1.267
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr21_+_30400229
|
1.260
|
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr1_+_64088886
|
1.255
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr1_-_173886401
|
1.215
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr4_-_84035888
|
1.211
|
|
PLAC8
|
placenta-specific 8
|
|
chr6_+_131894343
|
1.207
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr11_-_62996998
|
1.205
|
NM_199352
|
SLC22A25
|
solute carrier family 22, member 25
|
|
chr1_-_36948914
|
1.186
|
NM_000760
NM_156039
NM_172313
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte)
|
|
chr3_-_120401401
|
1.168
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr5_+_67586466
|
1.161
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr1_-_201081693
|
1.154
|
NM_000069
|
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit
|
|
chr22_+_26138110
|
1.150
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr6_-_128222175
|
1.144
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr11_-_118900078
|
1.141
|
NM_001164280
|
SLC37A4
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4
|
|
chr6_-_53530505
|
1.133
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr10_-_82049178
|
1.133
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr12_-_10151689
|
1.124
|
NM_001099431
NM_016509
|
CLEC1B
|
C-type lectin domain family 1, member B
|
|
chr20_+_32150139
|
1.118
|
NM_001039709
NM_005093
|
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2
|
|
chr1_-_150738292
|
1.113
|
NM_001199739
NM_004079
|
CTSS
|
cathepsin S
|
|
chr12_+_54891522
|
1.112
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr22_+_26138168
|
1.110
|
|
MYO18B
|
myosin XVIIIB
|
|
chr1_-_100643770
|
1.087
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chr2_+_182850550
|
1.084
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr1_-_157811491
|
1.080
|
NM_005894
|
CD5L
|
CD5 molecule-like
|
|
chr4_+_100495972
|
1.075
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr1_+_207262211
|
1.065
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr2_-_88285308
|
1.063
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr3_+_108541544
|
1.062
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr12_+_54891539
|
1.058
|
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr1_-_114414245
|
1.054
|
NM_001193431
NM_012411
NM_015967
|
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid)
|
|
chr1_+_40859016
|
1.050
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr9_+_137772653
|
1.043
|
NM_004108
NM_015837
|
FCN2
|
ficolin (collagen/fibrinogen domain containing lectin) 2 (hucolin)
|
|
chr11_+_46740769
|
1.043
|
|
F2
|
coagulation factor II (thrombin)
|
|
chr1_-_89641722
|
1.042
|
NM_207398
|
GBP7
|
guanylate binding protein 7
|
|
chr4_-_153273683
|
1.030
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr15_+_89631731
|
1.029
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr12_+_54891491
|
1.025
|
NM_005337
|
NCKAP1L
|
NCK-associated protein 1-like
|
|
chr15_+_89631695
|
1.024
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr4_-_84035908
|
1.023
|
NM_001130715
NM_016619
|
PLAC8
|
placenta-specific 8
|
|
chr7_-_14025826
|
1.017
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr3_+_133495925
|
1.004
|
|
TF
|
transferrin
|
|
chr16_+_7382750
|
1.000
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr19_-_54327428
|
0.999
|
NM_144687
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr2_-_165478357
|
0.997
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr15_+_89631625
|
0.996
|
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr11_+_112046207
|
0.983
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chrX_+_70765479
|
0.981
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr19_+_45445494
|
0.975
|
NM_001646
|
APOC4-APOC2
APOC4
|
APOC4-APOC2 readthrough
apolipoprotein C-IV
|
|
chr2_+_128176019
|
0.973
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr2_+_170366211
|
0.972
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr4_-_69536314
|
0.971
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr3_+_108855560
|
0.970
|
|
FLJ22763
|
uncharacterized LOC401081
|
|
chr15_+_86220267
|
0.959
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr16_-_21663962
|
0.957
|
NM_005849
|
IGSF6
|
immunoglobulin superfamily, member 6
|
|
chr2_+_172821874
|
0.953
|
|
HAT1
|
histone acetyltransferase 1
|
|
chr19_+_47421932
|
0.948
|
NM_004491
|
ARHGAP35
|
Rho GTPase activating protein 35
|
|
chr12_-_70093064
|
0.942
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr1_-_144997110
|
0.941
|
NM_001195260
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_-_13517525
|
0.941
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr15_+_52201590
|
0.935
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chrX_-_105282714
|
0.928
|
NM_000354
|
SERPINA7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7
|
|
chr16_+_31366529
|
0.928
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr14_+_24600674
|
0.927
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr16_+_8814567
|
0.924
|
NM_001127448
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr12_-_56756526
|
0.921
|
NM_001638
|
APOF
|
apolipoprotein F
|
|
chr8_-_72268720
|
0.919
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_+_115312777
|
0.918
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr10_-_96829074
|
0.918
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr12_-_10605928
|
0.917
|
NM_002259
NM_007328
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1
|
|
chr14_+_100485718
|
0.904
|
|
EVL
|
Enah/Vasp-like
|
|
chr9_+_136501484
|
0.903
|
NM_000787
|
DBH
|
dopamine beta-hydroxylase (dopamine beta-monooxygenase)
|
|
chr3_-_69171536
|
0.902
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr9_-_134585107
|
0.902
|
NM_198679
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1
|
|
chr4_-_100065372
|
0.901
|
NM_000670
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
chrX_-_63450486
|
0.901
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr19_+_55361897
|
0.897
|
NM_001242867
NM_006737
|
KIR3DL2
|
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 2
|
|
chr17_+_28707495
|
0.892
|
NM_001199775
|
CPD
|
carboxypeptidase D
|
|
chr1_-_160492982
|
0.892
|
NM_001184714
NM_001184715
NM_001184716
NM_052931
|
SLAMF6
|
SLAM family member 6
|
|
chr18_-_28742744
|
0.889
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr5_+_35856990
|
0.888
|
NM_002185
|
IL7R
|
interleukin 7 receptor
|
|
chr3_+_122044010
|
0.886
|
NM_005213
|
CSTA
|
cystatin A (stefin A)
|
|
chr7_-_126883568
|
0.878
|
NM_000845
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr5_+_53751430
|
0.877
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr3_+_188664939
|
0.873
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr4_-_155511838
|
0.873
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr15_+_58430394
|
0.869
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr3_-_52864687
|
0.864
|
NM_001166449
NM_002218
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4
|
|
chr12_-_111358353
|
0.861
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr2_-_21230572
|
0.861
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr1_-_11107150
|
0.860
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr3_+_63953419
|
0.858
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr14_-_38064299
|
0.849
|
|
FOXA1
|
forkhead box A1
|
|
chr4_+_95376396
|
0.848
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr5_-_96149804
|
0.848
|
NM_001198541
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1
|
|
chr2_+_53994928
|
0.844
|
NM_001008708
|
CHAC2
|
ChaC, cation transport regulator homolog 2 (E. coli)
|
|
chr15_-_42783293
|
0.843
|
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr9_+_127054567
|
0.839
|
NM_001166169
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr1_-_42384326
|
0.839
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr1_+_186265404
|
0.838
|
NM_001127708
NM_001127709
NM_001127710
NM_005807
|
PRG4
|
proteoglycan 4
|
|
chrX_-_124097601
|
0.836
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr17_-_10372875
|
0.835
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr12_-_7902068
|
0.829
|
NM_130441
NM_203503
|
CLEC4C
|
C-type lectin domain family 4, member C
|
|
chr10_+_28878666
|
0.826
|
|
WAC
|
WW domain containing adaptor with coiled-coil
|
|
chr3_-_138478184
|
0.825
|
NM_006219
|
PIK3CB
|
phosphoinositide-3-kinase, catalytic, beta polypeptide
|
|
chr17_-_72527512
|
0.825
|
|
CD300LB
|
CD300 molecule-like family member b
|
|
chr14_-_95608054
|
0.821
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr3_+_130569368
|
0.820
|
NM_001199180
NM_001199181
NM_001199182
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1
|
|
chr18_+_32556891
|
0.819
|
NM_001143826
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr2_-_32490688
|
0.819
|
NM_001199139
NM_021209
|
NLRC4
|
NLR family, CARD domain containing 4
|
|
chr7_-_36764000
|
0.817
|
NM_001177506
NM_001177507
NM_001637
|
AOAH
|
acyloxyacyl hydrolase (neutrophil)
|
|
chr9_-_116163550
|
0.816
|
|
ALAD
|
aminolevulinate dehydratase
|
|
chr9_+_96233420
|
0.815
|
|
FAM120A
|
family with sequence similarity 120A
|
|
chr16_+_31366468
|
0.812
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr16_+_6823809
|
0.807
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_173886464
|
0.807
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr5_+_54398473
|
0.803
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr12_-_10562744
|
0.800
|
NM_001199805
|
KLRC4-KLRK1
|
KLRC4-KLRK1 readthrough
|
|
chr16_+_31366519
|
0.799
|
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr4_+_156824846
|
0.798
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr14_+_70236266
|
0.796
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chrX_-_19817854
|
0.795
|
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr6_+_50681256
|
0.795
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|