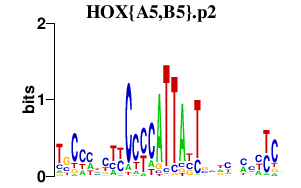
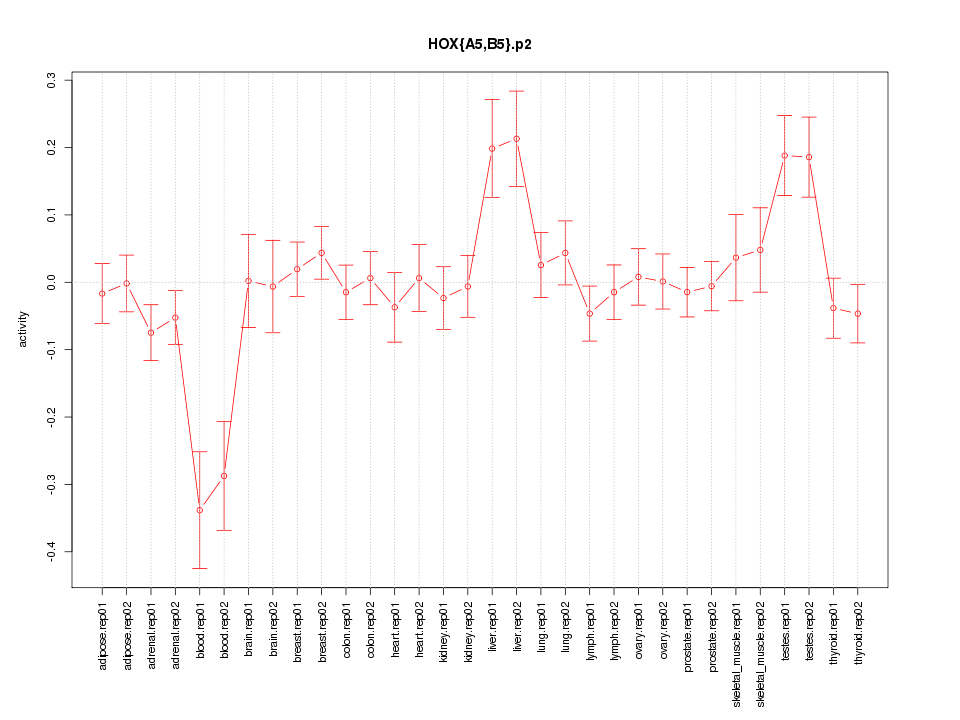
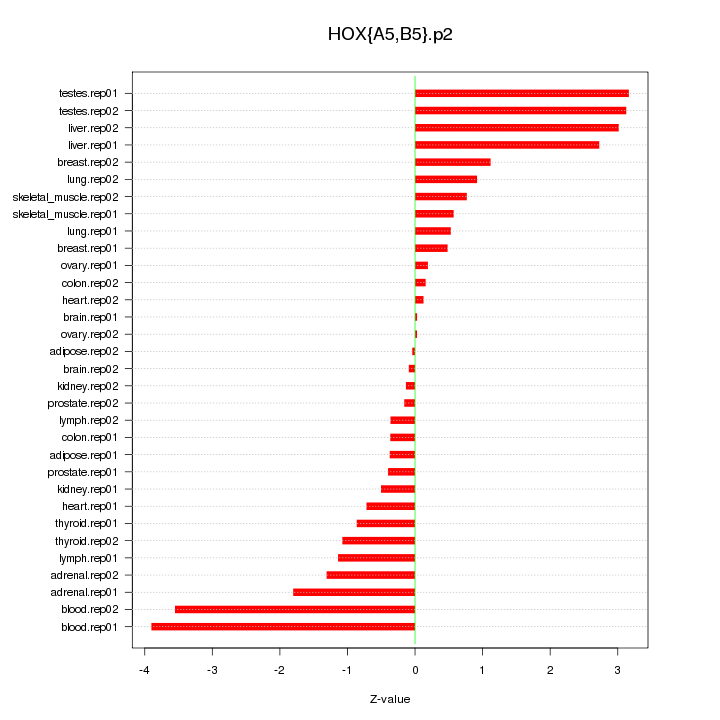
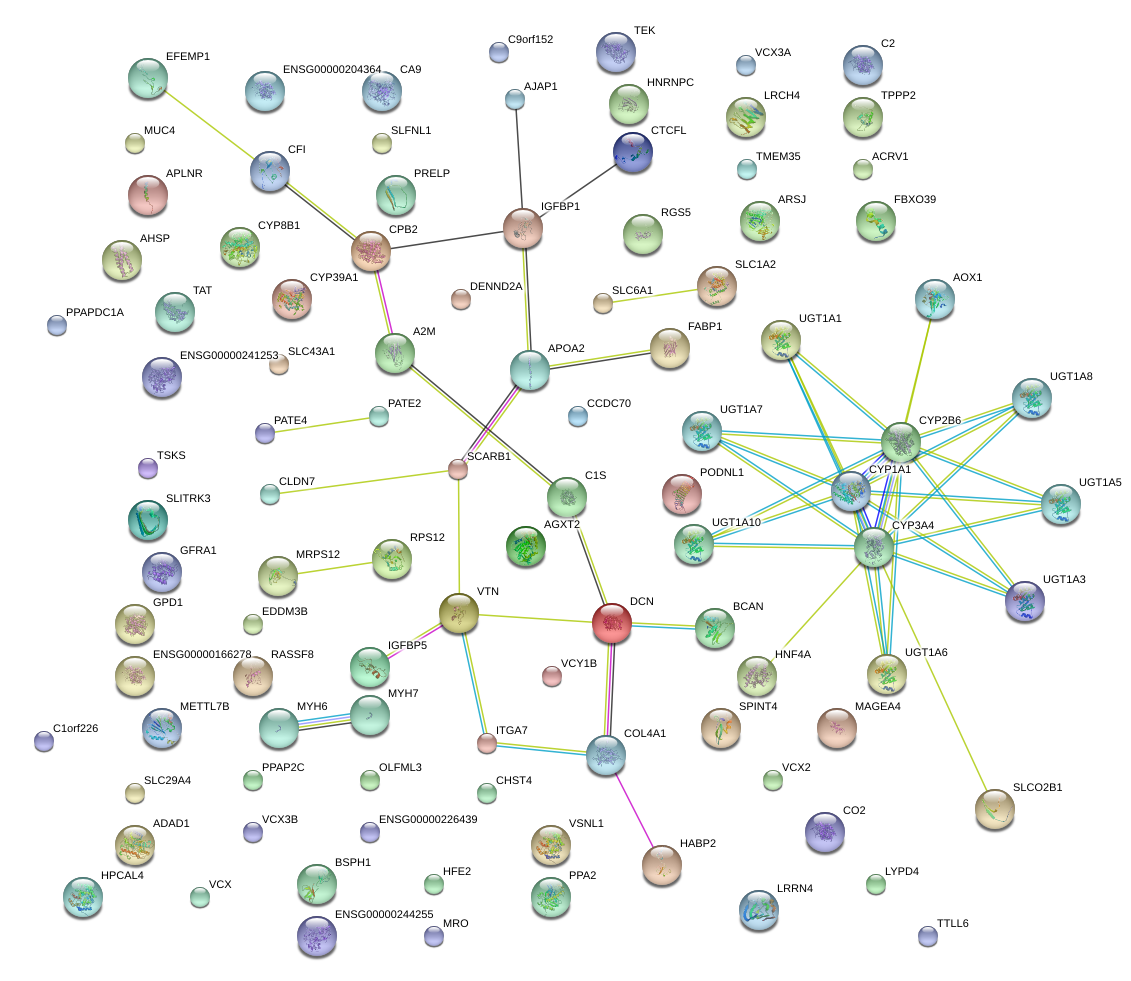
|
chr1_-_161193190
|
7.981
|
|
APOA2
|
apolipoprotein A-II
|
|
chr1_-_161193375
|
7.677
|
NM_001643
|
APOA2
|
apolipoprotein A-II
|
|
chr19_+_41515495
|
7.323
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr12_+_50497790
|
7.198
|
NM_005276
|
GPD1
|
glycerol-3-phosphate dehydrogenase 1 (soluble)
|
|
chr13_-_46679143
|
6.867
|
NM_001872
NM_016413
|
CPB2
|
carboxypeptidase B2 (plasma)
|
|
chr3_-_164914448
|
6.440
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr11_-_2018885
|
6.222
|
|
H19
RPS12
|
H19, imprinted maternally expressed transcript (non-protein coding)
ribosomal protein S12
|
|
chr7_-_99381703
|
6.220
|
NM_001202855
NM_017460
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4
|
|
chr2_-_88427535
|
6.138
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr11_-_2019007
|
5.822
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr10_+_104590430
|
5.395
|
|
|
|
|
chr7_+_5322553
|
5.068
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr12_-_9268513
|
5.067
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_+_56075329
|
4.989
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr14_+_21498344
|
4.921
|
NM_173846
|
TPPP2
|
tubulin polymerization-promoting protein family member 2
|
|
chr5_-_35048239
|
4.607
|
NM_031900
|
AGXT2
|
alanine--glyoxylate aminotransferase 2
|
|
chr12_-_91572328
|
4.536
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr20_+_42984088
|
4.535
|
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr2_+_234601511
|
4.490
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr16_-_71610948
|
4.479
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr1_-_163113129
|
4.468
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr3_-_42917632
|
4.367
|
NM_004391
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chrX_+_151084708
|
4.071
|
NM_002362
|
MAGEA4
|
melanoma antigen family A, 4
|
|
chr1_+_145413190
|
4.060
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr6_-_46620229
|
3.988
|
NM_016593
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1
|
|
chr5_+_133842276
|
3.975
|
|
|
|
|
chr3_-_195538699
|
3.974
|
NM_004532
NM_018406
NM_138297
|
MUC4
|
mucin 4, cell surface associated
|
|
chr1_+_203444882
|
3.970
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr11_-_2018446
|
3.969
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr12_+_56075418
|
3.741
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr11_+_74862031
|
3.704
|
NM_001145212
NM_007256
|
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1
|
|
chr14_-_23904849
|
3.614
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr19_-_42348507
|
3.461
|
NM_173506
|
LYPD4
|
LY6/PLAUR domain containing 4
|
|
chr12_-_56101465
|
3.423
|
|
ITGA7
|
integrin, alpha 7
|
|
chr10_+_115310589
|
3.392
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr11_-_2018334
|
3.388
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr1_+_114522029
|
3.340
|
NM_020190
|
OLFML3
|
olfactomedin-like 3
|
|
chr2_+_17721806
|
3.319
|
NM_003385
|
VSNL1
|
visinin-like 1
|
|
chrX_+_151086708
|
3.283
|
NM_001011550
|
MAGEA4
|
melanoma antigen family A, 4
|
|
chr15_-_75017710
|
3.281
|
NM_000499
|
CYP1A1
|
cytochrome P450, family 1, subfamily A, polypeptide 1
|
|
chr11_-_35440514
|
3.259
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_+_31868775
|
3.204
|
NM_001178063
|
C2
|
complement component 2
|
|
chrX_+_151085396
|
3.138
|
NM_001011549
|
MAGEA4
|
melanoma antigen family A, 4
|
|
chr19_-_48495339
|
3.058
|
NM_001128326
|
BSPH1
|
binder of sperm protein homolog 1
|
|
chr12_+_7167979
|
3.036
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr20_+_44350987
|
3.029
|
NM_178455
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4
|
|
chr4_-_114900802
|
3.028
|
NM_024590
|
ARSJ
|
arylsulfatase family, member J
|
|
chr14_+_21236585
|
2.929
|
NM_022360
|
EDDM3B
|
epididymal protein 3B
|
|
chr9_+_27109138
|
2.925
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr17_-_7166505
|
2.924
|
NM_001185022
|
CLDN7
|
claudin 7
|
|
chr10_+_122216465
|
2.918
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr1_+_203444955
|
2.895
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr13_-_110802708
|
2.876
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_+_201473942
|
2.871
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr17_-_46894380
|
2.870
|
NM_001130918
|
TTLL6
|
tubulin tyrosine ligase-like family, member 6
|
|
chr16_+_71560022
|
2.866
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr9_-_112970412
|
2.840
|
NM_001012993
|
C9orf152
|
chromosome 9 open reading frame 152
|
|
chr9_+_35673852
|
2.838
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr3_-_42917191
|
2.829
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr4_-_110723334
|
2.693
|
NM_000204
|
CFI
|
complement factor I
|
|
chr3_+_11034436
|
2.657
|
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr2_+_17721983
|
2.640
|
|
VSNL1
|
visinin-like 1
|
|
chr18_-_48346297
|
2.546
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr11_-_125648704
|
2.538
|
NM_212555
|
PATE2
|
prostate and testis expressed 2
|
|
chr12_-_125348346
|
2.532
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chrX_+_100333835
|
2.456
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr2_-_217540722
|
2.451
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr19_-_50266514
|
2.398
|
NM_021733
|
TSKS
|
testis-specific serine kinase substrate
|
|
chr17_+_6679551
|
2.395
|
NM_153230
|
FBXO39
|
F-box protein 39
|
|
chr11_-_2018568
|
2.375
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_-_291335
|
2.362
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr3_+_11034419
|
2.332
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr11_-_125550674
|
2.320
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr20_-_6034617
|
2.288
|
NM_152611
|
LRRN4
|
leucine rich repeat neuronal 4
|
|
chr16_+_31539202
|
2.258
|
NM_016633
|
AHSP
|
alpha hemoglobin stabilizing protein
|
|
chr11_-_57283145
|
2.232
|
NM_003627
|
SLC43A1
|
solute carrier family 43, member 1
|
|
chrX_-_6453158
|
2.217
|
NM_016379
|
VCX2
VCX3A
|
variable charge, X-linked 2
variable charge, X-linked 3A
|
|
chr2_-_56151273
|
2.190
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr7_-_140302311
|
2.153
|
NM_015689
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr4_+_123300594
|
2.148
|
NM_001159285
NM_001159295
|
ADAD1
|
adenosine deaminase domain containing 1 (testis-specific)
|
|
chr11_+_125703210
|
2.114
|
NM_001144874
|
PATE4
|
prostate and testis expressed 4
|
|
chr17_-_26697293
|
2.088
|
|
VTN
|
vitronectin
|
|
chr1_-_41487386
|
2.073
|
NM_001168247
NM_144990
|
SLFNL1
|
schlafen-like 1
|
|
chr7_+_79082910
|
2.046
|
|
MAGI2-AS3
|
MAGI2 antisense RNA 3 (non-protein coding)
|
|
chr1_+_162351519
|
2.046
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr12_+_26205490
|
2.015
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr20_-_56100138
|
2.006
|
NM_080618
|
CTCFL
|
CCCTC-binding factor (zinc finger protein)-like
|
|
chr10_-_118032975
|
2.000
|
NM_005264
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr7_+_45927958
|
1.983
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr1_+_4714668
|
1.981
|
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr11_-_57004674
|
1.977
|
NM_005161
|
APLNR
|
apelin receptor
|
|
chrX_+_8432870
|
1.940
|
NM_001001888
|
VCX3B
VCX3A
|
variable charge, X-linked 3B
variable charge, X-linked 3A
|
|
chr13_+_52436116
|
1.917
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr19_-_14064192
|
1.879
|
NM_001146254
NM_001146255
|
PODNL1
|
podocan-like 1
|
|
chr1_-_153958688
|
1.869
|
NM_002870
|
RAB13
|
RAB13, member RAS oncogene family
|
|
chrX_-_139047676
|
1.860
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr22_-_22901421
|
1.857
|
NM_206955
NM_206956
NM_006115
NM_206953
NM_206954
|
PRAME
|
preferentially expressed antigen in melanoma
|
|
chr12_-_6484557
|
1.853
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr12_-_125348422
|
1.834
|
NM_001082959
NM_005505
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr11_-_57089670
|
1.806
|
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chrX_+_51546154
|
1.802
|
NM_001005332
|
MAGED1
|
melanoma antigen family D, 1
|
|
chr15_+_82647307
|
1.784
|
|
UBE2Q2P2
|
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
|
|
chr4_-_100140330
|
1.751
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr7_-_98030165
|
1.750
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr2_-_74726665
|
1.747
|
|
LBX2
|
ladybird homeobox 2
|
|
chr8_+_7705401
|
1.744
|
NM_058203
NM_001081552
|
SPAG11B
SPAG11A
|
sperm associated antigen 11B
sperm associated antigen 11A
|
|
chr10_+_96522457
|
1.737
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr16_+_86544104
|
1.732
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr20_-_22564893
|
1.718
|
|
FOXA2
|
forkhead box A2
|
|
chr19_-_50143350
|
1.713
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr1_-_23810627
|
1.669
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chrY_+_26979966
|
1.669
|
NM_020364
NM_001005785
NM_001005786
NM_001005375
NM_020420
|
DAZ3
DAZ2
DAZ4
|
deleted in azoospermia 3
deleted in azoospermia 2
deleted in azoospermia 4
|
|
chrY_-_26959423
|
1.667
|
NM_020364
NM_020420
|
DAZ3
DAZ4
|
deleted in azoospermia 3
deleted in azoospermia 4
|
|
chr12_-_125348266
|
1.666
|
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr16_-_49860917
|
1.665
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr5_-_38595506
|
1.662
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr5_-_35938736
|
1.650
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr13_-_114538953
|
1.647
|
NM_001143945
|
GAS6
|
growth arrest-specific 6
|
|
chr17_-_26697224
|
1.632
|
|
VTN
|
vitronectin
|
|
chr3_-_170744458
|
1.627
|
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr11_-_18258315
|
1.624
|
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr2_-_216300444
|
1.611
|
|
FN1
|
fibronectin 1
|
|
chr7_+_100187195
|
1.611
|
NM_001163499
NM_012172
|
FBXO24
|
F-box protein 24
|
|
chrX_-_153847390
|
1.605
|
NM_001327
NM_139250
|
CTAG1B
CTAG1A
|
cancer/testis antigen 1B
cancer/testis antigen 1A
|
|
chrX_-_8769377
|
1.595
|
NM_001171186
NM_174951
|
FAM9A
|
family with sequence similarity 9, member A
|
|
chr8_-_93107442
|
1.591
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
1.584
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_44001177
|
1.580
|
NM_001193464
NM_015522
NM_016008
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr20_-_50418946
|
1.576
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chrX_+_105412297
|
1.571
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr17_-_73505914
|
1.571
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr6_+_121756744
|
1.569
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr19_-_55866063
|
1.566
|
NM_144613
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis)
|
|
chrX_-_38079980
|
1.530
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chrX_-_104465349
|
1.528
|
NM_031274
|
TEX13A
|
testis expressed 13A
|
|
chr17_-_26697310
|
1.526
|
|
VTN
|
vitronectin
|
|
chr1_+_16085245
|
1.517
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr12_-_9268475
|
1.493
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr17_+_38599651
|
1.487
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr11_-_2018703
|
1.485
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_+_48216600
|
1.483
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr16_+_2069520
|
1.478
|
NM_001099456
|
NPW
|
neuropeptide W
|
|
chr2_-_167350655
|
1.477
|
|
SCN7A
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr19_-_16008870
|
1.477
|
NM_001082
|
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2
|
|
chr13_-_39564857
|
1.456
|
NM_001144033
NM_145286
|
STOML3
|
stomatin (EPB72)-like 3
|
|
chr1_-_153599969
|
1.453
|
NM_001024211
|
S100A13
|
S100 calcium binding protein A13
|
|
chr6_+_31082526
|
1.452
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr10_-_94003002
|
1.445
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr19_+_51152634
|
1.430
|
NM_001195076
|
C19orf81
|
chromosome 19 open reading frame 81
|
|
chr6_-_33714681
|
1.427
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr14_-_21058880
|
1.427
|
NM_001024822
|
RNASE12
|
ribonuclease, RNase A family, 12 (non-active)
|
|
chr6_-_41006927
|
1.425
|
NM_173561
|
UNC5CL
|
unc-5 homolog C (C. elegans)-like
|
|
chrY_+_25365580
|
1.418
|
NM_020364
NM_001005785
NM_001005786
NM_020363
NM_020420
|
DAZ3
DAZ2
DAZ4
|
deleted in azoospermia 3
deleted in azoospermia 2
deleted in azoospermia 4
|
|
chr9_-_34710062
|
1.416
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr20_-_42816217
|
1.405
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chr13_+_58205943
|
1.402
|
|
PCDH17
|
protocadherin 17
|
|
chr1_+_169337193
|
1.398
|
NM_003666
|
BLZF1
|
basic leucine zipper nuclear factor 1
|
|
chrX_-_128657381
|
1.384
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr8_-_30706532
|
1.383
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr3_-_109056333
|
1.372
|
|
DPPA4
|
developmental pluripotency associated 4
|
|
chr17_+_7623038
|
1.371
|
NM_020877
|
DNAH2
|
dynein, axonemal, heavy chain 2
|
|
chrX_-_8139307
|
1.370
|
NM_016378
|
VCX2
|
variable charge, X-linked 2
|
|
chr6_+_161123224
|
1.369
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr4_+_22694547
|
1.368
|
NM_001128432
NM_020973
|
GBA3
|
glucosidase, beta, acid 3 (cytosolic)
|
|
chrX_+_37892786
|
1.367
|
NM_001163334
|
SYTL5
|
synaptotagmin-like 5
|
|
chr9_+_116172934
|
1.365
|
NM_152786
|
C9orf43
|
chromosome 9 open reading frame 43
|
|
chr19_-_49243836
|
1.353
|
NM_017805
|
RASIP1
|
Ras interacting protein 1
|
|
chrX_-_111326003
|
1.347
|
NM_012471
|
TRPC5
|
transient receptor potential cation channel, subfamily C, member 5
|
|
chr1_+_10490774
|
1.337
|
NM_001243768
|
APITD1-CORT
|
APITD1-CORT readthrough
|
|
chr1_-_153600714
|
1.334
|
NM_005979
|
S100A13
|
S100 calcium binding protein A13
|
|
chr13_+_29598746
|
1.331
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr12_-_6484904
|
1.322
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr12_+_54519836
|
1.321
|
|
LOC400043
|
uncharacterized LOC400043
|
|
chr19_-_291168
|
1.315
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr1_+_160085519
|
1.312
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chrX_+_114524291
|
1.307
|
NM_016383
|
LUZP4
|
leucine zipper protein 4
|
|
chr13_+_58205788
|
1.303
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr13_-_114018201
|
1.301
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr1_-_1310522
|
1.296
|
|
AURKAIP1
|
aurora kinase A interacting protein 1
|
|
chrX_-_119249774
|
1.285
|
NM_139282
|
RHOXF1
|
Rhox homeobox family, member 1
|
|
chr6_-_31981049
|
1.276
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr1_+_82266081
|
1.272
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr16_+_15489619
|
1.261
|
|
MPV17L
|
MPV17 mitochondrial membrane protein-like
|
|
chr3_-_58522873
|
1.257
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr17_+_4675175
|
1.257
|
NM_003963
|
TM4SF5
|
transmembrane 4 L six family member 5
|
|
chr11_+_71238312
|
1.234
|
NM_001012503
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr1_+_157963310
|
1.234
|
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr2_+_102770401
|
1.234
|
NM_000877
|
IL1R1
|
interleukin 1 receptor, type I
|
|
chr11_+_75428864
|
1.225
|
NM_025098
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chr7_+_99425635
|
1.217
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr20_+_42984338
|
1.216
|
NM_001030003
NM_001030004
NM_175914
|
HNF4A
|
hepatocyte nuclear factor 4, alpha
|
|
chr6_-_46922533
|
1.208
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr8_-_22785228
|
1.208
|
NM_144962
|
PEBP4
|
phosphatidylethanolamine-binding protein 4
|
|
chr20_-_22565050
|
1.207
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr15_+_82944821
|
1.207
|
|
AGSK1
|
golgin subfamily A member 2-like
|
|
chr22_+_25591072
|
1.202
|
|
KIAA1671
|
KIAA1671
|
|
chr9_-_33395265
|
1.192
|
|
AQP7
|
aquaporin 7
|
|
chr8_-_17752847
|
1.189
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr19_-_51845377
|
1.183
|
NM_001163922
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like
|
|
chr6_-_10419786
|
1.178
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|