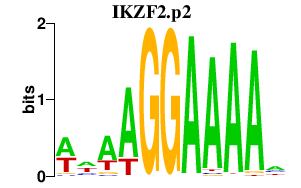
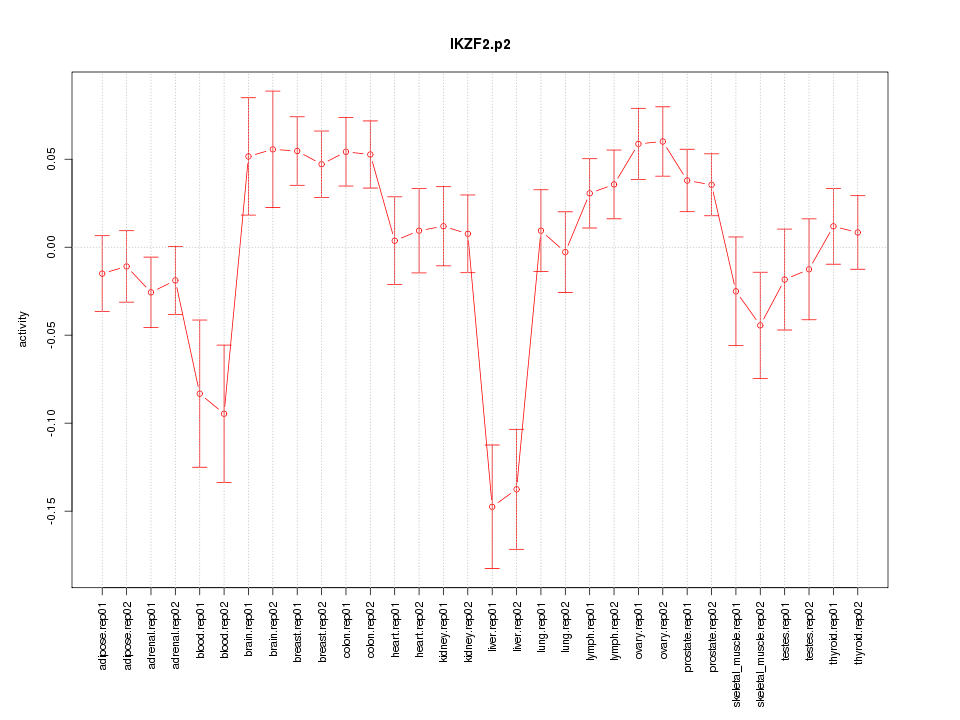
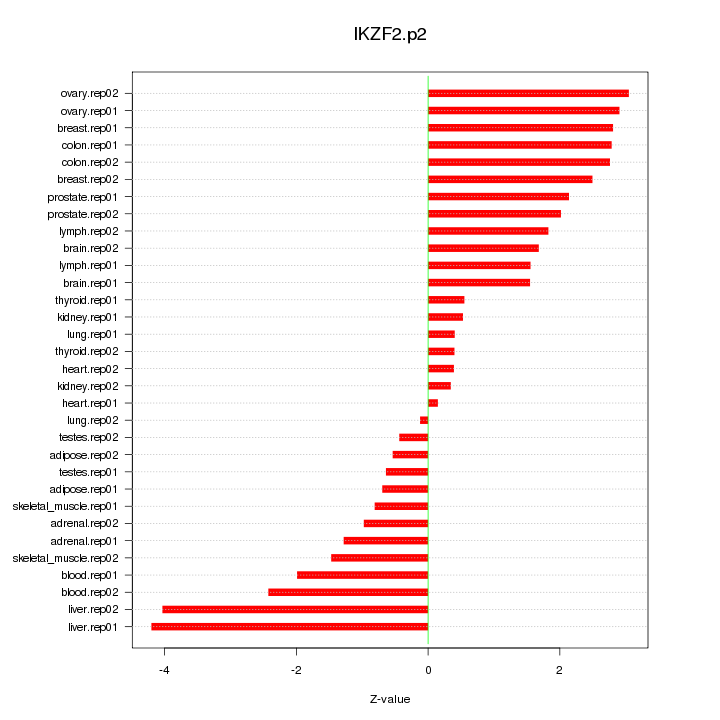
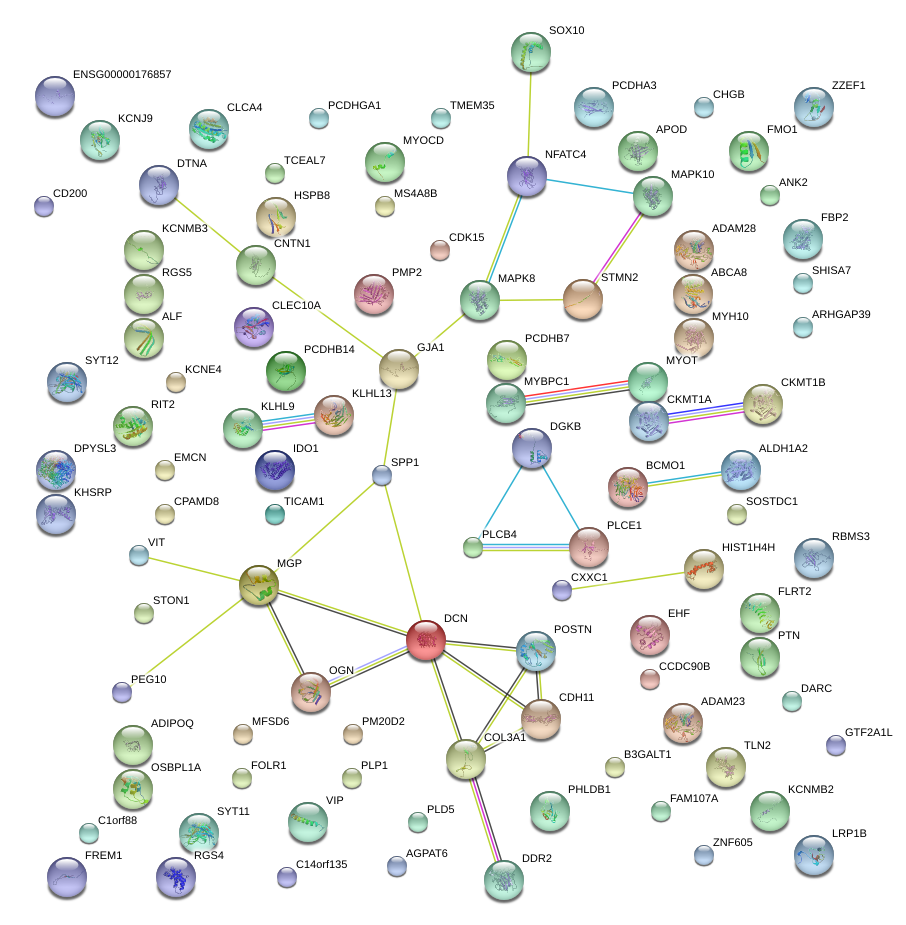
|
chr1_+_159175048
|
7.587
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr13_-_38172862
|
6.225
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr8_+_80523376
|
5.897
|
|
STMN2
|
stathmin-like 2
|
|
chr3_-_195310770
|
5.458
|
|
APOD
|
apolipoprotein D
|
|
chr7_-_16505292
|
5.424
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chrX_+_102585113
|
4.992
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr15_+_43885251
|
4.776
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr3_-_195310985
|
4.751
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr6_+_153071931
|
4.651
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr17_-_66951381
|
4.643
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_+_112051914
|
4.507
|
NM_001004196
NM_005944
|
CD200
|
CD200 molecule
|
|
chr11_+_60467046
|
4.263
|
NM_031457
|
MS4A8B
|
membrane-spanning 4-domains, subfamily A, member 8B
|
|
chr22_-_38380538
|
4.224
|
NM_006941
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chrX_+_100333835
|
3.967
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr7_+_94297448
|
3.934
|
|
PEG10
|
paternally expressed 10
|
|
chr1_+_171217632
|
3.818
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr7_-_14880954
|
3.767
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr4_+_88896868
|
3.732
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr17_+_12623605
|
3.670
|
NM_001146313
|
MYOCD
|
myocardin
|
|
chr2_+_168675181
|
3.581
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr4_+_88896838
|
3.552
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr20_+_5891973
|
3.495
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr20_+_9198036
|
3.407
|
NM_182797
|
PLCB4
|
phospholipase C, beta 4
|
|
chr22_-_38379868
|
3.381
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr5_+_140602914
|
3.325
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr1_+_162602198
|
3.318
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr7_-_137028533
|
3.264
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr16_+_81272293
|
3.183
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr3_-_12586962
|
3.180
|
NM_001195279
|
LOC100129480
|
uncharacterized LOC100129480
|
|
chr3_+_178276487
|
3.171
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr1_+_155853546
|
3.128
|
|
SYT11
|
synaptotagmin XI
|
|
chr1_+_111889194
|
3.013
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr1_-_163113938
|
2.996
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr12_-_15038724
|
2.987
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr3_+_186560478
|
2.976
|
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr8_+_39771327
|
2.928
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr18_+_32290195
|
2.913
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr14_+_85996512
|
2.891
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr5_+_140552201
|
2.821
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr3_+_186560462
|
2.774
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr3_-_58563070
|
2.739
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr18_+_32335939
|
2.718
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_+_207406815
|
2.675
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr2_-_142889269
|
2.665
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr11_+_34654010
|
2.655
|
NM_001206616
|
EHF
|
ets homologous factor
|
|
chr3_+_29322802
|
2.648
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr4_-_101439071
|
2.641
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chrX_+_103031797
|
2.623
|
|
PLP1
|
proteolipid protein 1
|
|
chr4_-_87281374
|
2.607
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr17_-_8424217
|
2.562
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr5_-_87969135
|
2.558
|
|
LINC00461
|
long intergenic non-protein coding RNA 461
|
|
chr4_+_88896801
|
2.555
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr1_-_242612757
|
2.543
|
NM_001195811
NM_001195812
|
PLD5
|
phospholipase D family, member 5
|
|
chr5_+_137203559
|
2.538
|
|
MYOT
|
myotilin
|
|
chr8_-_145838887
|
2.523
|
NM_025251
|
ARHGAP39
|
Rho GTPase activating protein 39
|
|
chr3_-_58563490
|
2.515
|
NM_001076778
NM_007177
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chrX_+_103031753
|
2.508
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr4_-_87281177
|
2.500
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr18_-_40695483
|
2.496
|
NM_002930
|
RIT2
|
Ras-like without CAAX 2
|
|
chrX_-_117119282
|
2.478
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr4_+_88896899
|
2.470
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_191301020
|
2.449
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr1_+_163038934
|
2.447
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr12_+_119616594
|
2.433
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr12_-_91573264
|
2.432
|
NM_133503
|
DCN
|
decorin
|
|
chr15_+_43985083
|
2.432
|
NM_001015001
|
CKMT1A
|
creatine kinase, mitochondrial 1A
|
|
chr15_+_63050761
|
2.412
|
|
TLN2
|
talin 2
|
|
chr11_+_71903172
|
2.405
|
NM_016729
|
FOLR1
|
folate receptor 1 (adult)
|
|
chrX_+_103031433
|
2.401
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr1_+_87012758
|
2.397
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr15_-_58306185
|
2.393
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr6_+_121756744
|
2.372
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr2_+_36923902
|
2.359
|
|
VIT
|
vitrin
|
|
chr7_-_137028433
|
2.358
|
|
PTN
|
pleiotrophin
|
|
chr2_+_223916861
|
2.344
|
NM_080671
|
KCNE4
|
potassium voltage-gated channel, Isk-related family, member 4
|
|
chr7_-_137028275
|
2.329
|
|
PTN
|
pleiotrophin
|
|
chr14_+_24837225
|
2.325
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr2_+_48796158
|
2.321
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr5_+_140180782
|
2.312
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr4_+_113739203
|
2.310
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr5_-_146833150
|
2.304
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr9_-_95166827
|
2.295
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr2_+_189839113
|
2.295
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_202671197
|
2.294
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr7_-_137028370
|
2.291
|
|
PTN
|
pleiotrophin
|
|
chr2_+_189839078
|
2.287
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr8_-_82359620
|
2.282
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr16_-_65155965
|
2.269
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr12_+_101988746
|
2.264
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr9_-_14910992
|
2.250
|
NM_144966
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr11_+_118477212
|
2.248
|
NM_015157
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr1_+_160051343
|
2.246
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr12_+_41086189
|
2.243
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr9_-_97356007
|
2.239
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr10_+_95848811
|
2.231
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr5_+_140710203
|
2.225
|
NM_018912
NM_031993
|
PCDHGA1
|
protocadherin gamma subfamily A, 1
|
|
chr3_+_112052020
|
2.225
|
|
CD200
|
CD200 molecule
|
|
chrX_+_100805415
|
2.222
|
NM_016608
|
ARMCX1
|
armadillo repeat containing, X-linked 1
|
|
chr9_-_93405066
|
2.217
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr4_+_134070443
|
2.203
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr1_-_79472360
|
2.201
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr11_+_101983170
|
2.195
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr5_+_140718353
|
2.190
|
NM_018915
NM_032009
|
PCDHGA2
|
protocadherin gamma subfamily A, 2
|
|
chr5_-_92916924
|
2.185
|
|
FLJ42709
|
uncharacterized LOC441094
|
|
chr18_-_64271215
|
2.183
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr2_+_189839132
|
2.175
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr12_-_18243052
|
2.164
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr5_+_140174443
|
2.134
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr1_+_162602308
|
2.130
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr17_+_48262816
|
2.122
|
|
|
|
|
chr15_+_74466171
|
2.114
|
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr22_-_38379790
|
2.112
|
|
SOX10
|
SRY (sex determining region Y)-box 10
|
|
chr2_-_217560247
|
2.112
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr7_+_153584383
|
2.110
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr5_+_137203544
|
2.108
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr3_+_12392993
|
2.091
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr10_-_7682741
|
2.076
|
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr12_+_26348268
|
2.067
|
NM_001135823
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr17_-_29624249
|
2.065
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr4_-_120243257
|
2.062
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chrX_+_56755666
|
2.056
|
|
LOC550643
|
uncharacterized LOC550643
|
|
chr8_+_75896707
|
2.054
|
NM_031461
|
CRISPLD1
|
cysteine-rich secretory protein LCCL domain containing 1
|
|
chr3_-_123135188
|
2.053
|
NM_001199642
|
ADCY5
|
adenylate cyclase 5
|
|
chrX_-_6145887
|
2.048
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr7_+_134576187
|
2.030
|
|
CALD1
|
caldesmon 1
|
|
chr1_+_163038564
|
2.026
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr3_-_112358510
|
2.013
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr3_-_161089747
|
2.010
|
NM_001040100
|
SPTSSB
|
serine palmitoyltransferase, small subunit B
|
|
chr17_-_42908178
|
2.002
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr11_+_101918168
|
1.991
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr7_+_94024201
|
1.964
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chrX_-_133119475
|
1.944
|
|
GPC3
|
glypican 3
|
|
chr5_-_96478502
|
1.937
|
NM_153234
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr19_-_42931500
|
1.932
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr6_-_47009992
|
1.932
|
NM_025048
NM_153840
|
GPR110
|
G protein-coupled receptor 110
|
|
chr10_+_63422718
|
1.931
|
NM_173554
|
C10orf107
|
chromosome 10 open reading frame 107
|
|
chr5_-_146460991
|
1.923
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr11_+_62475165
|
1.912
|
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr8_-_95274520
|
1.910
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr5_+_155754172
|
1.905
|
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr16_-_66952729
|
1.899
|
NM_001204744
NM_001204745
NM_001204746
NM_004062
|
CDH16
|
cadherin 16, KSP-cadherin
|
|
chr4_-_87374282
|
1.895
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr2_-_238322770
|
1.891
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr4_+_124317884
|
1.866
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr3_+_238274
|
1.866
|
NM_001253387
NM_006614
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr6_+_123100860
|
1.852
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr5_+_140625146
|
1.850
|
NM_018935
|
PCDHB15
|
protocadherin beta 15
|
|
chr16_-_65155984
|
1.846
|
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr2_+_167744996
|
1.844
|
NM_001079810
NM_001199143
NM_152381
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr7_+_142829169
|
1.843
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr2_-_238322806
|
1.819
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr17_+_34391639
|
1.809
|
NM_002988
|
CCL18
|
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated)
|
|
chr5_+_80529138
|
1.802
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chr8_-_95274540
|
1.802
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr2_+_74120132
|
1.795
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr4_-_138453628
|
1.795
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr3_-_93747453
|
1.787
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr12_+_78225068
|
1.787
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chrX_-_133119624
|
1.786
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr8_-_6420454
|
1.785
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr3_+_361365
|
1.773
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr5_+_173472692
|
1.770
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr12_-_91505215
|
1.769
|
|
LUM
|
lumican
|
|
chr3_-_116164363
|
1.768
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr5_-_96478438
|
1.768
|
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr3_+_115342302
|
1.766
|
|
GAP43
|
growth associated protein 43
|
|
chr8_+_24771276
|
1.764
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_-_71551564
|
1.763
|
|
TSPAN8
|
tetraspanin 8
|
|
chr5_+_140247767
|
1.758
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr4_-_159093523
|
1.758
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr1_+_100111430
|
1.753
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr1_+_101185195
|
1.746
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr2_+_27301693
|
1.742
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr8_-_108510094
|
1.738
|
NM_001146
NM_001199859
|
ANGPT1
|
angiopoietin 1
|
|
chr5_-_142077508
|
1.730
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr13_+_23755059
|
1.724
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chrX_+_135251454
|
1.722
|
NM_001159704
|
FHL1
|
four and a half LIM domains 1
|
|
chr8_+_17433941
|
1.720
|
NM_006207
|
PDGFRL
|
platelet-derived growth factor receptor-like
|
|
chr2_-_217540722
|
1.714
|
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr21_-_27945267
|
1.710
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr2_-_238322814
|
1.708
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_203320185
|
1.705
|
|
FMOD
|
fibromodulin
|
|
chr12_+_175872
|
1.704
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr1_+_171154387
|
1.703
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr11_-_118047145
|
1.685
|
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr12_+_26348488
|
1.680
|
NM_005086
|
SSPN
|
sarcospan (Kras oncogene-associated gene)
|
|
chr7_-_83824216
|
1.676
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr4_+_151503076
|
1.672
|
NM_006439
|
MAB21L2
|
mab-21-like 2 (C. elegans)
|
|
chr18_-_3874688
|
1.671
|
NM_001242762
NM_001242763
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr11_-_40314510
|
1.664
|
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr11_-_68780667
|
1.657
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr5_+_92918924
|
1.651
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr7_+_86273219
|
1.645
|
NM_000840
|
GRM3
|
glutamate receptor, metabotropic 3
|
|
chr16_-_65155881
|
1.642
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr12_-_91576579
|
1.636
|
|
DCN
|
decorin
|
|
chr8_-_22089517
|
1.634
|
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein
|
|
chr4_+_55095433
|
1.633
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr11_+_62475113
|
1.632
|
NM_012202
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3
|
|
chr2_+_27301434
|
1.627
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr1_-_36565849
|
1.619
|
NM_005202
|
COL8A2
|
collagen, type VIII, alpha 2
|