|
chr21_+_47401644
|
4.489
|
NM_001848
|
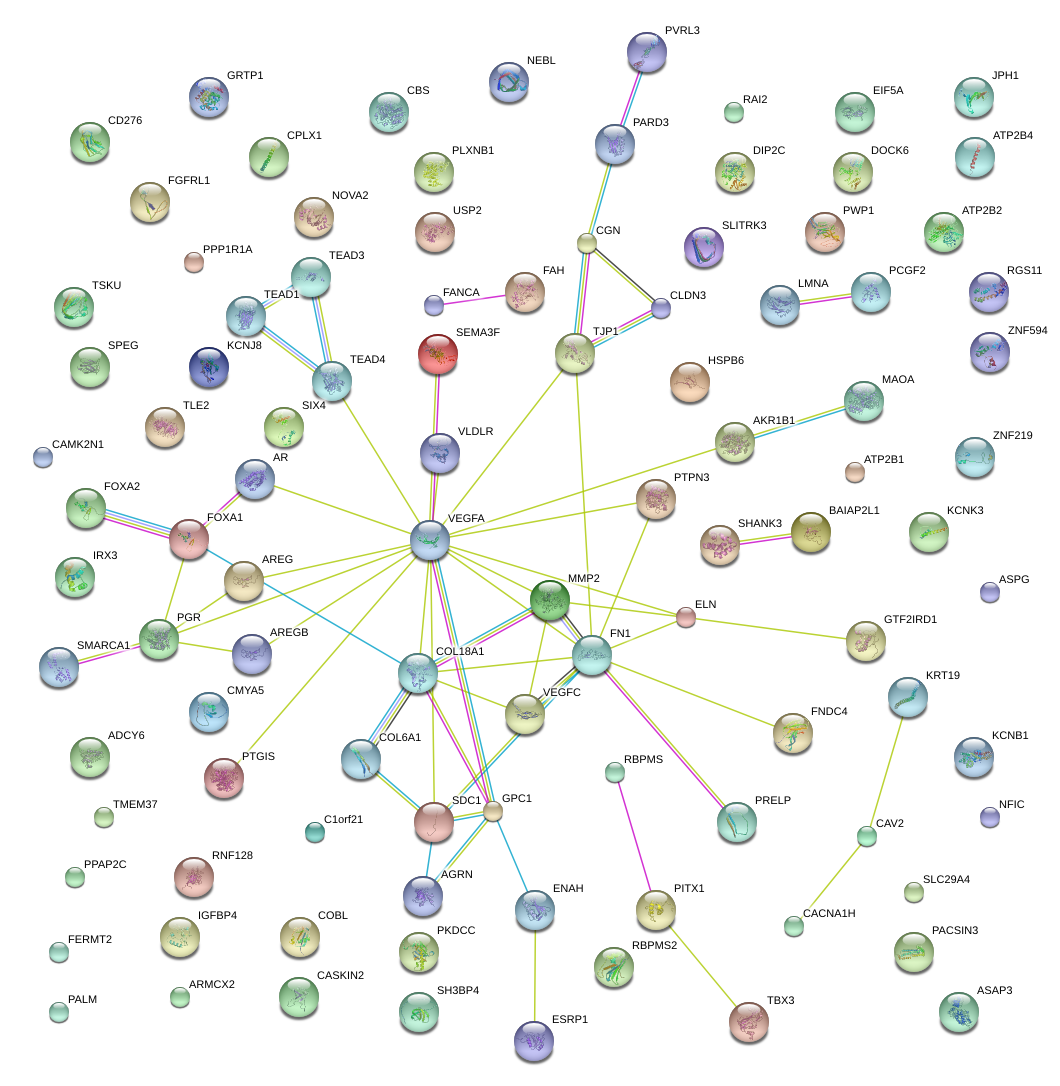
COL6A1
|
collagen, type VI, alpha 1
|
|
chr12_-_21927614
|
3.838
|
NM_004982
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr13_-_114018365
|
3.771
|
NM_024719
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr19_-_291335
|
3.411
|
NM_003712
NM_177526
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr4_-_819879
|
3.058
|
NM_006651
|
CPLX1
|
complexin 1
|
|
chr4_+_1005609
|
3.025
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr22_+_51113069
|
2.851
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr2_+_42275017
|
2.708
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr7_-_51384458
|
2.703
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr12_-_54982271
|
2.698
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr13_-_114018201
|
2.685
|
|
GRTP1
|
growth hormone regulated TBC protein 1
|
|
chr12_+_3068442
|
2.644
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr7_+_73868397
|
2.533
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr15_+_80445129
|
2.520
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr15_-_65067696
|
2.499
|
NM_194272
|
RBPMS2
|
RNA binding protein with multiple splicing 2
|
|
chrX_+_43514157
|
2.487
|
|
MAOA
|
monoamine oxidase A
|
|
chr12_-_115121939
|
2.321
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr6_-_35464716
|
2.120
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr7_-_51384331
|
1.964
|
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr11_+_76494132
|
1.957
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr20_-_22564893
|
1.903
|
|
FOXA2
|
forkhead box A2
|
|
chr14_-_53417636
|
1.890
|
|
FERMT2
|
fermitin family member 2
|
|
chr21_-_44495894
|
1.875
|
NM_000071
|
CBS
|
cystathionine-beta-synthase
|
|
chr1_-_23810627
|
1.842
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr4_+_1005395
|
1.813
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr14_-_21566497
|
1.800
|
|
ZNF219
|
zinc finger protein 219
|
|
chr1_+_203444882
|
1.757
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr7_+_73868236
|
1.736
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr19_+_3359138
|
1.733
|
NM_001245005
NM_205843
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr19_+_708766
|
1.690
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr15_+_73976720
|
1.680
|
|
CD276
|
CD276 molecule
|
|
chr2_-_20424627
|
1.667
|
NM_002997
|
SDC1
|
syndecan 1
|
|
chr15_+_80445177
|
1.656
|
NM_000137
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr7_-_98030165
|
1.651
|
|
BAIAP2L1
|
BAI1-associated protein 2-like 1
|
|
chr3_+_50192420
|
1.644
|
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr16_+_1203240
|
1.643
|
NM_001005407
NM_021098
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|
|
chr10_-_35103894
|
1.640
|
NM_001184785
NM_001184786
NM_001184787
NM_001184788
NM_001184789
NM_001184790
NM_001184791
NM_001184792
NM_001184793
NM_001184794
NM_019619
|
PARD3
|
par-3 partitioning defective 3 homolog (C. elegans)
|
|
chr6_+_43738757
|
1.637
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr8_+_30241943
|
1.624
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr5_-_134369841
|
1.617
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr1_+_151483861
|
1.554
|
NM_020770
|
CGN
|
cingulin
|
|
chr19_-_291168
|
1.510
|
NM_177543
|
PPAP2C
|
phosphatidic acid phosphatase type 2C
|
|
chr14_+_104552015
|
1.509
|
NM_001080464
|
ASPG
|
asparaginase homolog (S. cerevisiae)
|
|
chr17_-_73511453
|
1.506
|
|
CASKIN2
|
CASK interacting protein 2
|
|
chr2_+_26915390
|
1.503
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr14_-_38064176
|
1.488
|
|
FOXA1
|
forkhead box A1
|
|
chr2_-_216300444
|
1.483
|
|
FN1
|
fibronectin 1
|
|
chr19_-_36246411
|
1.465
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr16_-_325867
|
1.463
|
NM_003834
NM_183337
|
RGS11
|
regulator of G-protein signaling 11
|
|
chrX_-_128657381
|
1.463
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr3_-_48470715
|
1.457
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr20_-_48184652
|
1.440
|
NM_000961
|
PTGIS
|
prostaglandin I2 (prostacyclin) synthase
|
|
chr14_-_38064440
|
1.439
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr1_+_156052330
|
1.432
|
|
LMNA
|
lamin A/C
|
|
chr17_-_39684499
|
1.425
|
|
KRT19
|
keratin 19
|
|
chr16_-_54320367
|
1.420
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr7_+_73868111
|
1.418
|
NM_001199207
NM_005685
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr14_-_61190458
|
1.415
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr12_-_49182718
|
1.409
|
NM_020983
|
ADCY6
|
adenylate cyclase 6
|
|
chr3_+_110790460
|
1.402
|
NM_001243286
NM_015480
|
PVRL3
|
poliovirus receptor-related 3
|
|
chr19_-_36246206
|
1.400
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr10_-_735522
|
1.377
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr4_-_177713664
|
1.363
|
NM_005429
|
VEGFC
|
vascular endothelial growth factor C
|
|
chrX_+_66763868
|
1.359
|
NM_000044
|
AR
|
androgen receptor
|
|
chr11_-_47207425
|
1.358
|
NM_001184975
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr1_+_184356364
|
1.357
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr7_+_73442486
|
1.339
|
|
ELN
|
elastin
|
|
chr15_+_73976777
|
1.337
|
|
CD276
|
CD276 molecule
|
|
chrX_-_128657453
|
1.332
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr1_-_20812727
|
1.330
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr21_-_44496423
|
1.312
|
NM_001178008
NM_001178009
|
CBS
|
cystathionine-beta-synthase
|
|
chr2_+_120189342
|
1.306
|
NM_183240
|
TMEM37
|
transmembrane protein 37
|
|
chr9_-_112260529
|
1.296
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr14_-_21566728
|
1.292
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr1_+_203444955
|
1.290
|
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr8_+_95653399
|
1.289
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr17_-_73511626
|
1.288
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr17_+_7210929
|
1.286
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chrX_+_105969875
|
1.267
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr2_+_235860616
|
1.259
|
NM_014521
|
SH3BP4
|
SH3-domain binding protein 4
|
|
chr7_+_116139619
|
1.250
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr20_-_48099178
|
1.248
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr11_-_119234859
|
1.234
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr19_-_3028901
|
1.233
|
NM_003260
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr1_-_225840660
|
1.223
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr2_+_241375235
|
1.221
|
|
GPC1
|
glypican 1
|
|
chr7_+_5322553
|
1.218
|
NM_001040661
NM_153247
|
SLC29A4
|
solute carrier family 29 (nucleoside transporters), member 4
|
|
chr8_+_95653272
|
1.203
|
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr10_-_21462987
|
1.199
|
|
NEBL
|
nebulette
|
|
chr5_+_78985645
|
1.192
|
NM_153610
|
CMYA5
|
cardiomyopathy associated 5
|
|
chrX_-_17879355
|
1.181
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr2_-_27718059
|
1.177
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr17_-_36904545
|
1.173
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr17_+_38599651
|
1.169
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr1_+_955609
|
1.169
|
|
AGRN
|
agrin
|
|
chr10_-_21463019
|
1.163
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr16_+_55512801
|
1.160
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr11_-_47207938
|
1.156
|
NM_001184974
NM_016223
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr8_-_75233461
|
1.156
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr19_-_46476556
|
1.155
|
NM_002516
|
NOVA2
|
neuro-oncological ventral antigen 2
|
|
chr14_-_53417808
|
1.152
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr2_+_241375030
|
1.151
|
NM_002081
|
GPC1
|
glypican 1
|
|
chr21_+_46825001
|
1.147
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr1_-_20812271
|
1.144
|
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr3_-_164914448
|
1.133
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr3_-_10547267
|
1.127
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr4_+_1006251
|
1.126
|
NM_021923
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr15_-_30114536
|
1.120
|
NM_003257
NM_175610
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr7_-_73184535
|
1.108
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr1_-_23810721
|
1.107
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr9_+_2622134
|
1.100
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr15_+_73976689
|
1.100
|
|
CD276
|
CD276 molecule
|
|
chr19_-_11373117
|
1.098
|
NM_020812
|
DOCK6
|
dedicator of cytokinesis 6
|
|
chr14_-_38064299
|
1.097
|
|
FOXA1
|
forkhead box A1
|
|
chr2_+_220325533
|
1.097
|
NM_001173476
|
SPEG
|
SPEG complex locus
|
|
chr11_+_12695851
|
1.094
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr11_+_113930296
|
1.091
|
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr8_-_10587942
|
1.087
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr17_-_40828441
|
1.083
|
NM_024927
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3
|
|
chr11_+_113930430
|
1.082
|
NM_006006
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr9_+_2622052
|
1.082
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chrX_+_135229536
|
1.081
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr2_-_216300770
|
1.076
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr15_+_80445340
|
1.075
|
|
FAH
|
fumarylacetoacetate hydrolase (fumarylacetoacetase)
|
|
chr7_+_73442426
|
1.075
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr20_-_60942300
|
1.071
|
NM_005560
|
LAMA5
|
laminin, alpha 5
|
|
chr3_+_184097860
|
1.070
|
NM_003741
|
CHRD
|
chordin
|
|
chr6_+_132129195
|
1.067
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr10_-_118031542
|
1.067
|
NM_145793
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr9_+_2622099
|
1.051
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr11_-_65640199
|
1.046
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chr9_+_133320247
|
1.043
|
|
ASS1
|
argininosuccinate synthase 1
|
|
chr8_+_95653340
|
1.041
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr9_+_133320093
|
1.040
|
NM_000050
NM_054012
|
ASS1
|
argininosuccinate synthase 1
|
|
chr7_-_140340575
|
1.034
|
|
DENND2A
|
DENN/MADD domain containing 2A
|
|
chr1_+_3371020
|
1.033
|
NM_014448
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16
|
|
chr2_-_110371669
|
1.029
|
NM_144710
NM_178584
|
SEPT10
|
septin 10
|
|
chr6_-_33714681
|
1.028
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr1_+_164528586
|
1.026
|
NM_001204961
NM_001204963
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr7_+_94023872
|
1.019
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr8_-_144923019
|
1.018
|
NM_178564
|
NRBP2
|
nuclear receptor binding protein 2
|
|
chr6_+_132129154
|
1.018
|
NM_006208
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr12_+_3186520
|
1.008
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr9_+_133971902
|
1.007
|
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr2_-_227664330
|
1.004
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr9_-_140444835
|
1.001
|
NM_001098537
NM_152286
|
PNPLA7
|
patatin-like phospholipase domain containing 7
|
|
chr5_+_149980627
|
0.997
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr17_-_74533623
|
0.996
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chrX_-_128657440
|
0.993
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr2_+_220306772
|
0.993
|
|
SPEG
|
SPEG complex locus
|
|
chr3_-_169381417
|
0.990
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr4_-_16900256
|
0.988
|
|
LDB2
|
LIM domain binding 2
|
|
chr13_-_114538953
|
0.988
|
NM_001143945
|
GAS6
|
growth arrest-specific 6
|
|
chr13_+_98795710
|
0.979
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr3_-_149375584
|
0.979
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr5_-_178772430
|
0.978
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr6_+_19837599
|
0.977
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr4_+_3294711
|
0.975
|
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr4_-_16900028
|
0.975
|
|
LDB2
|
LIM domain binding 2
|
|
chr6_+_1390068
|
0.975
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr6_+_17393610
|
0.971
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr11_-_68780667
|
0.970
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr2_+_217498082
|
0.967
|
NM_000597
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chr7_+_94024201
|
0.965
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr1_+_955468
|
0.959
|
NM_198576
|
AGRN
|
agrin
|
|
chr3_+_110790774
|
0.957
|
NM_001243288
|
PVRL3
|
poliovirus receptor-related 3
|
|
chrX_-_133119624
|
0.956
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr17_-_1389013
|
0.947
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr6_+_133562488
|
0.946
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr9_+_133971862
|
0.945
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr9_+_140446308
|
0.937
|
NM_032477
|
MRPL41
|
mitochondrial ribosomal protein L41
|
|
chr22_+_29279573
|
0.936
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr18_+_46065367
|
0.935
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr21_-_44846927
|
0.928
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr3_+_50192847
|
0.928
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr16_-_52580805
|
0.925
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr17_+_47653474
|
0.923
|
|
NXPH3
|
neurexophilin 3
|
|
chr16_-_65155881
|
0.918
|
NM_001797
|
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast)
|
|
chr10_-_99790584
|
0.917
|
NM_001206528
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr1_+_184356186
|
0.914
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr11_-_2159784
|
0.911
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr4_-_16900348
|
0.909
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr5_+_42423811
|
0.908
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr18_+_19749403
|
0.906
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr22_-_36424397
|
0.905
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_-_106532447
|
0.904
|
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr11_+_111783476
|
0.901
|
|
HSPB2-C11orf52
HSPB2
|
HSPB2-C11orf52 readthrough (non-protein coding)
heat shock 27kDa protein 2
|
|
chr17_-_78009453
|
0.899
|
NM_019020
|
TBC1D16
|
TBC1 domain family, member 16
|
|
chr12_+_6875686
|
0.898
|
|
PTMS
|
parathymosin
|
|
chr17_+_7211260
|
0.897
|
NM_001143761
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr5_+_176513819
|
0.897
|
NM_002011
NM_213647
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr11_-_57092283
|
0.891
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr1_+_184356227
|
0.890
|
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr10_-_44880452
|
0.888
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr11_+_57365577
|
0.883
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr5_+_2752261
|
0.881
|
NM_178569
|
C5orf38
|
chromosome 5 open reading frame 38
|
|
chr1_-_201368434
|
0.881
|
|
LAD1
|
ladinin 1
|
|
chr12_-_118541655
|
0.879
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr22_+_45898690
|
0.864
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr9_-_34589679
|
0.859
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|