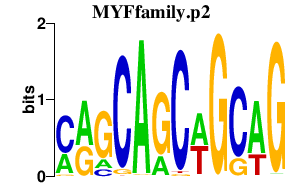
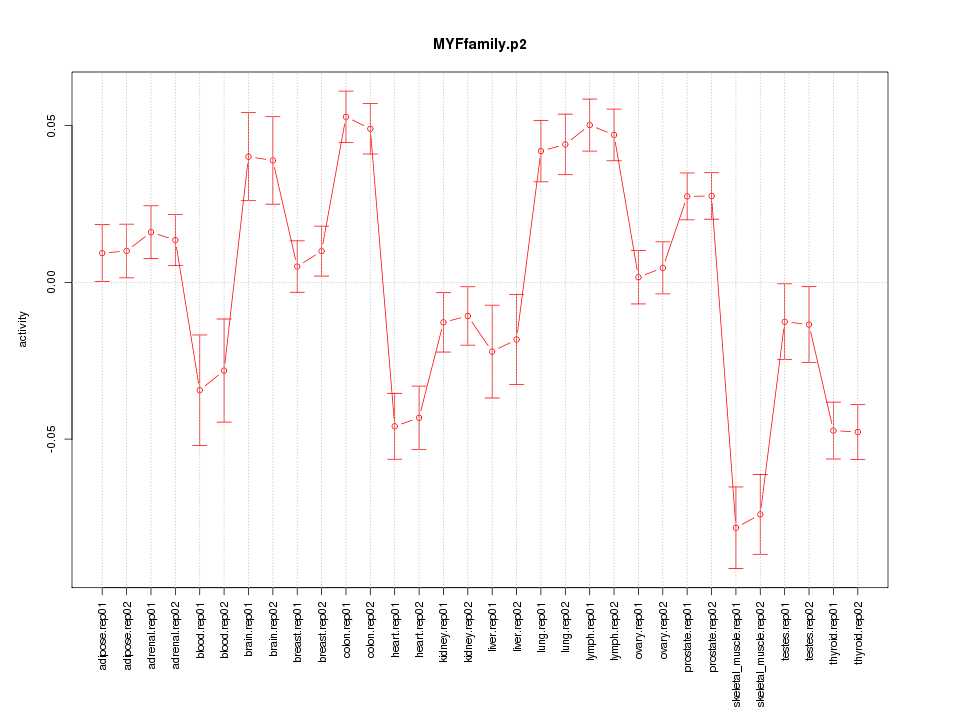
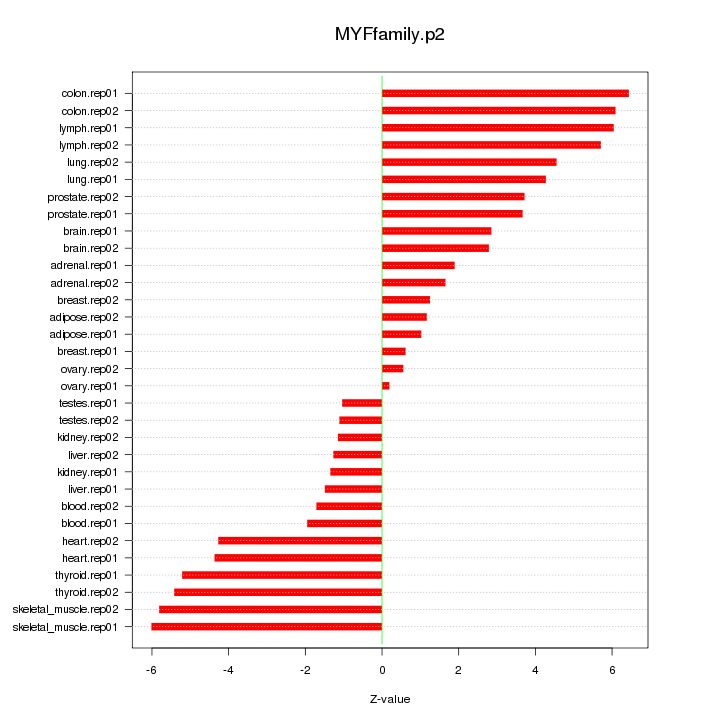
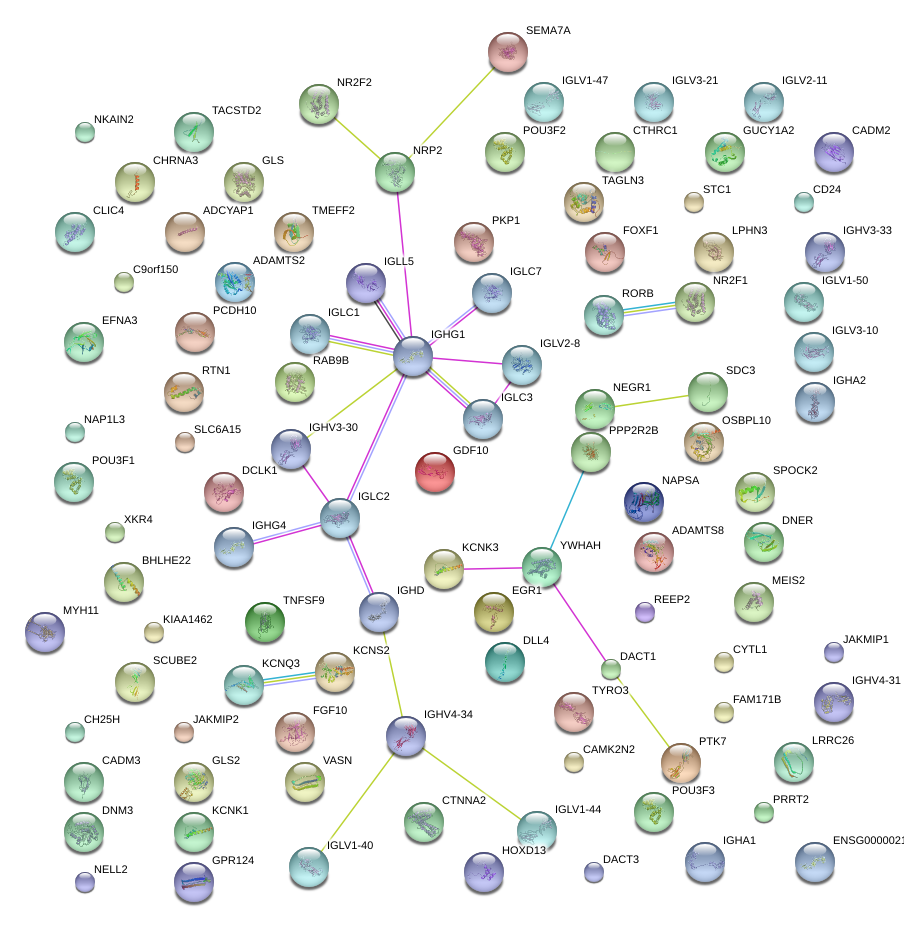
|
chr11_-_130298502
|
9.433
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr11_-_130298305
|
8.752
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr3_+_159943628
|
8.251
|
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr3_+_111717585
|
6.666
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr2_+_187558654
|
6.475
|
NM_177454
|
FAM171B
|
family with sequence similarity 171, member B
|
|
chr3_+_159943252
|
6.146
|
NM_001168214
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr8_+_65493610
|
6.039
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr4_-_5021095
|
6.024
|
NM_018659
|
CYTL1
|
cytokine-like 1
|
|
chr8_-_23712297
|
5.461
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr22_+_22712091
|
5.438
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr3_+_111718006
|
5.437
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr11_-_9113092
|
5.414
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr16_+_29823557
|
5.371
|
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr16_+_86544233
|
5.366
|
|
FOXF1
|
forkhead box F1
|
|
chr5_+_92920592
|
5.307
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_+_79740059
|
5.256
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr2_-_230579198
|
4.947
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr6_+_99282597
|
4.794
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr5_+_137801178
|
4.725
|
NM_001964
|
EGR1
|
early growth response 1
|
|
chr1_-_59042827
|
4.651
|
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr2_+_105471745
|
4.328
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr3_+_85008031
|
4.326
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr5_-_138725544
|
4.231
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr5_-_146258143
|
4.190
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr8_-_133493003
|
4.173
|
NM_004519
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr5_+_137774632
|
4.170
|
NM_016606
|
REEP2
|
receptor accessory protein 2
|
|
chr1_+_155051263
|
4.137
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr10_-_90967048
|
4.060
|
NM_003956
|
CH25H
|
cholesterol 25-hydroxylase
|
|
chr1_+_233749749
|
4.022
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr14_-_107083398
|
3.989
|
|
IGH@
IGHA1
IGHG1
|
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_106641585
|
3.975
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_86544104
|
3.961
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr9_+_12775713
|
3.933
|
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr15_+_96873845
|
3.810
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr3_-_183979074
|
3.793
|
NM_033259
|
CAMK2N2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2
|
|
chr1_-_72748147
|
3.738
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr16_-_15835382
|
3.643
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr8_+_104383785
|
3.608
|
NM_138455
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr1_+_171810611
|
3.578
|
NM_001136127
NM_015569
|
DNM3
|
dynamin 3
|
|
chr8_+_104383701
|
3.550
|
|
CTHRC1
|
collagen triple helix repeat containing 1
|
|
chr15_-_78913119
|
3.516
|
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3
|
|
chr10_-_73848309
|
3.475
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr14_-_106780535
|
3.464
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_60337350
|
3.436
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr15_-_37391596
|
3.431
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr14_-_106774311
|
3.342
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr15_-_74726258
|
3.311
|
NM_001146029
NM_003612
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr1_+_25071906
|
3.310
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr5_+_137774897
|
3.302
|
|
REEP2
|
receptor accessory protein 2
|
|
chr10_-_48438975
|
3.294
|
NM_004962
|
GDF10
|
growth differentiation factor 10
|
|
chr4_+_134073180
|
3.238
|
|
PCDH10
|
protocadherin 10
|
|
chr22_+_22676814
|
3.213
|
|
|
|
|
chr5_-_147162056
|
3.200
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr16_+_4430878
|
3.169
|
|
VASN
|
vasorin
|
|
chr2_+_176957448
|
3.163
|
NM_000523
|
HOXD13
|
homeobox D13
|
|
chr2_+_191745568
|
3.152
|
|
GLS
|
glutaminase
|
|
chr2_+_191745547
|
3.140
|
|
GLS
|
glutaminase
|
|
chr6_+_124125068
|
3.078
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chrX_-_92928566
|
3.064
|
NM_004538
|
NAP1L3
|
nucleosome assembly protein 1-like 3
|
|
chr1_+_159141376
|
3.034
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr1_+_25071759
|
2.997
|
NM_013943
|
CLIC4
|
chloride intracellular channel 4
|
|
chr1_-_72748491
|
2.990
|
|
|
|
|
chr10_-_73848530
|
2.990
|
NM_001244950
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr4_-_6202251
|
2.978
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr2_+_79740171
|
2.970
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr2_+_206547131
|
2.959
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr5_+_137801166
|
2.945
|
|
EGR1
|
early growth response 1
|
|
chr10_-_30316338
|
2.932
|
|
KIAA1462
|
KIAA1462
|
|
chr6_+_43043944
|
2.886
|
NM_002821
NM_152880
NM_152881
NM_152882
|
PTK7
|
PTK7 protein tyrosine kinase 7
|
|
chr8_+_37654757
|
2.879
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr19_-_50868877
|
2.878
|
NM_004851
|
NAPSA
|
napsin A aspartic peptidase
|
|
chr4_+_62066945
|
2.839
|
|
LPHN3
|
latrophilin 3
|
|
chr3_-_32022734
|
2.839
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr15_+_41851153
|
2.784
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr2_+_26915390
|
2.734
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr5_-_178772430
|
2.727
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr12_-_85306579
|
2.726
|
NM_001146335
NM_018057
NM_182767
|
SLC6A15
|
solute carrier family 6 (neutral amino acid transporter), member 15
|
|
chr15_+_41221530
|
2.700
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr1_+_25071967
|
2.664
|
|
CLIC4
|
chloride intracellular channel 4
|
|
chr1_+_201252579
|
2.663
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr9_-_140064426
|
2.647
|
NM_001013653
|
LRRC26
|
leucine rich repeat containing 26
|
|
chr13_-_36705432
|
2.622
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr11_-_106888829
|
2.608
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr2_+_191745791
|
2.607
|
|
GLS
|
glutaminase
|
|
chr8_+_99439249
|
2.600
|
NM_020697
|
KCNS2
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2
|
|
chr14_+_59104736
|
2.593
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr18_+_904943
|
2.582
|
NM_001099733
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr5_-_147162251
|
2.562
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr16_+_29823408
|
2.559
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chrY_-_21154560
|
2.557
|
NM_013230
|
CD24
CD24P4
|
CD24 molecule
CD24 molecule pseudogene 4
|
|
chr5_-_44388666
|
2.535
|
NM_004465
|
FGF10
|
fibroblast growth factor 10
|
|
chrX_-_103087106
|
2.534
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr1_-_31381436
|
2.530
|
NM_014654
|
SDC3
|
syndecan 3
|
|
chr9_+_77112251
|
2.502
|
NM_006914
|
RORB
|
RAR-related orphan receptor B
|
|
chr8_+_56015013
|
2.452
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr19_+_6531009
|
2.447
|
NM_003811
|
TNFSF9
|
tumor necrosis factor (ligand) superfamily, member 9
|
|
chr22_+_22550112
|
2.444
|
|
|
|
|
chr2_+_206547352
|
2.437
|
|
NRP2
|
neuropilin 2
|
|
chr22_+_32340435
|
2.424
|
NM_003405
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr14_-_106453162
|
2.417
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_-_193059599
|
2.417
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr1_-_67390413
|
2.412
|
NM_024763
NM_207014
|
WDR78
|
WD repeat domain 78
|
|
chr7_-_131241321
|
2.412
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr12_+_79258517
|
2.407
|
|
SYT1
|
synaptotagmin I
|
|
chr22_+_23229959
|
2.396
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr3_-_189838907
|
2.393
|
NM_018192
|
LEPREL1
|
leprecan-like 1
|
|
chr10_-_73848278
|
2.372
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr3_+_139654001
|
2.366
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr8_+_37654395
|
2.354
|
NM_032777
|
GPR124
|
G protein-coupled receptor 124
|
|
chr16_+_23847271
|
2.341
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr4_-_57976550
|
2.331
|
NM_001253835
NM_001553
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr2_+_128403438
|
2.310
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr8_+_24771276
|
2.297
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr6_+_149721494
|
2.290
|
NM_001002255
|
SUMO4
|
SMT3 suppressor of mif two 3 homolog 4 (S. cerevisiae)
|
|
chr6_+_7727010
|
2.275
|
NM_001718
|
BMP6
|
bone morphogenetic protein 6
|
|
chr5_-_178772328
|
2.268
|
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr12_-_6579754
|
2.264
|
NM_014231
NM_016830
NM_199245
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1)
|
|
chr19_-_47137938
|
2.245
|
NM_033258
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8
|
|
chr1_-_58716173
|
2.242
|
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr9_-_140063854
|
2.237
|
|
|
|
|
chr18_-_22931970
|
2.231
|
|
ZNF521
|
zinc finger protein 521
|
|
chr15_+_48413168
|
2.224
|
NM_205850
|
SLC24A5
|
solute carrier family 24, member 5
|
|
chrX_-_99665270
|
2.219
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr3_+_139654206
|
2.217
|
|
CLSTN2
|
calsyntenin 2
|
|
chr22_+_23010757
|
2.216
|
|
|
|
|
chr7_+_119913688
|
2.212
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr12_+_49688908
|
2.209
|
NM_006262
|
PRPH
|
peripherin
|
|
chr22_+_22550159
|
2.201
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr19_+_41699076
|
2.198
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr12_+_121570630
|
2.194
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr10_+_85954390
|
2.184
|
NM_001171971
NM_033100
|
CDHR1
|
cadherin-related family member 1
|
|
chr20_+_62328068
|
2.173
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr2_+_191745546
|
2.165
|
NM_014905
|
GLS
|
glutaminase
|
|
chr20_-_31071187
|
2.157
|
NM_080616
|
C20orf112
|
chromosome 20 open reading frame 112
|
|
chr1_+_153651050
|
2.150
|
NM_000906
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A)
|
|
chr21_+_22370622
|
2.141
|
NM_004540
|
NCAM2
|
neural cell adhesion molecule 2
|
|
chr10_-_15130773
|
2.139
|
NM_001039844
|
ACBD7
|
acyl-CoA binding domain containing 7
|
|
chr8_+_24771268
|
2.137
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr12_-_40499604
|
2.135
|
NM_052885
|
SLC2A13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr4_-_57976516
|
2.133
|
|
IGFBP7
|
insulin-like growth factor binding protein 7
|
|
chr22_+_42372763
|
2.126
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr20_+_2673193
|
2.126
|
NM_001110514
|
EBF4
|
early B-cell factor 4
|
|
chr2_-_219925237
|
2.120
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr7_+_153749740
|
2.115
|
NM_130797
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr22_+_32340592
|
2.106
|
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr16_+_4838397
|
2.091
|
NM_001253790
NM_001253791
|
LOC440335
|
uncharacterized LOC440335
|
|
chr3_-_195306274
|
2.083
|
|
APOD
|
apolipoprotein D
|
|
chr22_+_22764094
|
2.082
|
|
|
|
|
chr1_-_59043165
|
2.075
|
NM_002353
|
TACSTD2
|
tumor-associated calcium signal transducer 2
|
|
chr2_+_219824346
|
2.072
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr12_+_4918341
|
2.071
|
NM_002235
|
KCNA6
|
potassium voltage-gated channel, shaker-related subfamily, member 6
|
|
chr15_+_89182038
|
2.052
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr8_+_37654833
|
2.045
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr4_-_13546037
|
2.043
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr8_+_37654406
|
2.043
|
|
GPR124
|
G protein-coupled receptor 124
|
|
chr12_-_53901371
|
2.040
|
NM_003717
|
NPFF
|
neuropeptide FF-amide peptide precursor
|
|
chr10_+_25464289
|
2.023
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr15_-_48470453
|
2.018
|
NM_016132
|
MYEF2
|
myelin expression factor 2
|
|
chr18_+_21594349
|
2.017
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr6_-_31514620
|
2.013
|
NM_001204078
NM_130463
NM_138282
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr19_+_17638064
|
2.002
|
|
FAM129C
|
family with sequence similarity 129, member C
|
|
chr7_-_11871735
|
1.997
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr15_-_74726000
|
1.990
|
NM_001146030
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group)
|
|
chr1_+_163041694
|
1.982
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr16_+_2867227
|
1.973
|
|
PRSS21
|
protease, serine, 21 (testisin)
|
|
chr14_-_106452693
|
1.971
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr11_-_117747331
|
1.968
|
|
FXYD6
|
FXYD domain containing ion transport regulator 6
|
|
chr18_+_905296
|
1.967
|
NM_001117
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary)
|
|
chr4_-_168155732
|
1.966
|
NM_001040159
NM_001204352
NM_001204354
NM_001204355
NM_001204356
NM_001204358
NM_001251967
NM_016950
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr7_-_37956377
|
1.960
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr16_-_29910340
|
1.950
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr2_-_47797469
|
1.947
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr4_-_25864430
|
1.941
|
NM_015187
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr4_-_168155691
|
1.933
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chrX_-_38079980
|
1.930
|
NM_001170750
NM_001170751
NM_001170752
NM_006307
|
SRPX
|
sushi-repeat containing protein, X-linked
|
|
chr20_+_62328002
|
1.928
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr19_+_3224700
|
1.921
|
NM_021938
NM_001172673
|
CELF5
|
CUGBP, Elav-like family member 5
|
|
chr20_-_3662635
|
1.915
|
NM_025220
NM_153202
|
ADAM33
|
ADAM metallopeptidase domain 33
|
|
chr1_+_233749917
|
1.914
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr7_-_27239724
|
1.914
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr18_+_21269528
|
1.913
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr6_+_14117857
|
1.910
|
NM_001040280
NM_004233
|
CD83
|
CD83 molecule
|
|
chr5_+_140346289
|
1.910
|
NM_031883
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr20_-_35374468
|
1.909
|
|
NDRG3
|
NDRG family member 3
|
|
chrX_+_23352984
|
1.907
|
NM_173495
|
PTCHD1
|
patched domain containing 1
|
|
chr9_-_139940556
|
1.900
|
|
NPDC1
|
neural proliferation, differentiation and control, 1
|
|
chr6_+_151186814
|
1.896
|
NM_001242767
NM_001242769
NM_015440
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr19_-_7701852
|
1.894
|
|
|
|
|
chr3_-_58652469
|
1.892
|
NM_138805
|
FAM3D
|
family with sequence similarity 3, member D
|
|
chr3_-_4508955
|
1.887
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr2_+_158114109
|
1.882
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr1_-_58716207
|
1.876
|
NM_021080
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr6_-_31514393
|
1.876
|
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2
|
|
chr12_+_86268072
|
1.875
|
NM_006183
|
NTS
|
neurotensin
|
|
chr14_-_23821423
|
1.874
|
NM_020372
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr9_+_87284625
|
1.873
|
NM_001007097
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr4_+_166128823
|
1.873
|
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr19_-_18314731
|
1.870
|
NM_002866
|
RAB3A
|
RAB3A, member RAS oncogene family
|
|
chr8_-_140715234
|
1.863
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr2_+_115199898
|
1.862
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|