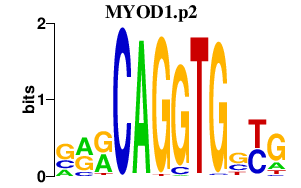
|
chr19_-_11591439
|
4.970
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr9_+_17578952
|
3.822
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr3_+_50712671
|
3.618
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr2_+_79740059
|
3.617
|
NM_001164883
NM_004389
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr20_-_62130390
|
3.466
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr16_+_67679058
|
3.125
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr3_+_35681008
|
3.123
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chrX_-_13956530
|
2.983
|
|
GPM6B
|
glycoprotein M6B
|
|
chrX_-_13956445
|
2.939
|
|
|
|
|
chr2_+_79740171
|
2.837
|
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2
|
|
chr10_+_99332197
|
2.807
|
NM_001129981
NM_020349
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle)
|
|
chr1_+_205012329
|
2.762
|
NM_005076
|
CNTN2
|
contactin 2 (axonal)
|
|
chr6_+_96463844
|
2.760
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr16_+_23847271
|
2.748
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr5_-_149669187
|
2.740
|
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chrX_-_13956644
|
2.730
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr5_-_149669400
|
2.693
|
NM_015981
NM_171825
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr10_+_88428263
|
2.591
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr16_+_67679011
|
2.499
|
NM_001013838
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr15_+_75118950
|
2.478
|
NM_001030005
|
CPLX3
|
complexin 3
|
|
chr16_+_23847359
|
2.469
|
|
PRKCB
|
protein kinase C, beta
|
|
chr1_+_236849753
|
2.439
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr10_+_122216465
|
2.380
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr17_-_42992852
|
2.368
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr20_-_1974701
|
2.299
|
NM_001190898
NM_001190899
NM_024411
|
PDYN
|
prodynorphin
|
|
chr1_-_201346783
|
2.283
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr7_-_124405029
|
2.267
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr16_+_56623266
|
2.234
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr11_-_35441099
|
2.229
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr9_-_101470836
|
2.172
|
NM_005458
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2
|
|
chr14_-_77608132
|
2.145
|
NM_174976
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr10_-_16564003
|
2.112
|
NM_001010908
|
C1QL3
|
complement component 1, q subcomponent-like 3
|
|
chr1_-_201346804
|
2.071
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr11_+_73358593
|
2.018
|
NM_001130034
NM_001130035
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr16_+_24857551
|
2.015
|
NM_052944
|
SLC5A11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11
|
|
chr19_+_54385429
|
2.012
|
NM_002739
|
PRKCG
|
protein kinase C, gamma
|
|
chr2_+_228029280
|
1.984
|
NM_000091
|
COL4A3
|
collagen, type IV, alpha 3 (Goodpasture antigen)
|
|
chr18_-_24445652
|
1.976
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr2_-_230579198
|
1.949
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr12_-_70093064
|
1.936
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr18_-_74691951
|
1.913
|
|
MBP
|
myelin basic protein
|
|
chr19_+_54926604
|
1.909
|
NM_001005367
NM_001201461
NM_020659
|
TTYH1
|
tweety homolog 1 (Drosophila)
|
|
chr5_+_173472692
|
1.909
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr16_+_25703346
|
1.908
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr5_+_176237435
|
1.858
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr2_-_220118610
|
1.857
|
NM_006000
|
TUBA4A
|
tubulin, alpha 4a
|
|
chr9_-_139948448
|
1.836
|
NM_001246
NM_203468
|
ENTPD2
|
ectonucleoside triphosphate diphosphohydrolase 2
|
|
chr11_-_12030128
|
1.832
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_+_12132063
|
1.795
|
NM_014632
|
MICAL2
|
microtubule associated monoxygenase, calponin and LIM domain containing 2
|
|
chr8_-_143695812
|
1.784
|
NM_015193
|
ARC
|
activity-regulated cytoskeleton-associated protein
|
|
chr19_-_7939325
|
1.773
|
NM_001190467
|
FLJ22184
|
putative uncharacterized protein FLJ22184
|
|
chr14_+_67999915
|
1.766
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr5_+_173472737
|
1.764
|
|
HMP19
|
HMP19 protein
|
|
chr19_+_36359346
|
1.756
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr4_-_46995440
|
1.751
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chrX_-_102565805
|
1.750
|
NM_001168399
NM_001168400
NM_001168401
NM_032621
|
BEX2
|
brain expressed X-linked 2
|
|
chr5_-_11904127
|
1.747
|
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr9_+_138594030
|
1.733
|
NM_020822
|
KCNT1
|
potassium channel, subfamily T, member 1
|
|
chr3_+_85008031
|
1.730
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chrX_+_152224765
|
1.727
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr1_+_1981902
|
1.699
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr2_-_241759627
|
1.696
|
NM_001244008
NM_004321
|
KIF1A
|
kinesin family member 1A
|
|
chr3_-_118753428
|
1.681
|
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr10_-_21462987
|
1.675
|
|
NEBL
|
nebulette
|
|
chr10_-_56560947
|
1.668
|
NM_001142763
NM_001142764
NM_001142765
NM_001142766
NM_001142767
NM_001142768
NM_001142769
NM_001142770
NM_001142771
NM_001142772
NM_001142773
NM_033056
|
PCDH15
|
protocadherin-related 15
|
|
chr5_+_175223519
|
1.659
|
NM_006650
|
CPLX2
|
complexin 2
|
|
chr14_-_77607958
|
1.643
|
|
ZDHHC22
|
zinc finger, DHHC-type containing 22
|
|
chr19_-_6502224
|
1.633
|
NM_006087
|
TUBB4A
|
tubulin, beta 4A class IVa
|
|
chr9_-_91793375
|
1.632
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr2_-_70780621
|
1.624
|
|
TGFA
|
transforming growth factor, alpha
|
|
chr2_-_175629140
|
1.608
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr6_+_150464156
|
1.603
|
NM_030949
|
PPP1R14C
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C
|
|
chr10_-_21463019
|
1.601
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr3_-_118753665
|
1.592
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr12_-_109458844
|
1.590
|
NM_018711
|
SVOP
|
SV2 related protein homolog (rat)
|
|
chr16_-_31439650
|
1.586
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr6_+_34433849
|
1.584
|
NM_020804
|
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr14_-_65289812
|
1.582
|
NM_000347
NM_001024858
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr9_+_140918042
|
1.576
|
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr18_-_74844762
|
1.565
|
NM_001025100
NM_001025101
|
MBP
|
myelin basic protein
|
|
chr11_-_111782447
|
1.553
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr1_+_153600859
|
1.546
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr1_-_40782924
|
1.543
|
NM_001852
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr16_-_30042123
|
1.543
|
NM_031478
|
FAM57B
|
family with sequence similarity 57, member B
|
|
chr16_+_330605
|
1.538
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr3_-_183543295
|
1.534
|
NM_024871
|
MAP6D1
|
MAP6 domain containing 1
|
|
chr14_+_29236882
|
1.527
|
|
FOXG1
|
forkhead box G1
|
|
chrX_+_152224862
|
1.523
|
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr1_+_169075869
|
1.520
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr1_-_51796230
|
1.512
|
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr1_-_40782981
|
1.496
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr20_+_61273823
|
1.496
|
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr11_+_22214574
|
1.492
|
NM_001142649
NM_213599
|
ANO5
|
anoctamin 5
|
|
chr16_+_58498216
|
1.480
|
|
NDRG4
|
NDRG family member 4
|
|
chr17_-_57184034
|
1.478
|
|
TRIM37
|
tripartite motif containing 37
|
|
chr9_-_93405066
|
1.449
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr16_+_58498182
|
1.428
|
|
NDRG4
|
NDRG family member 4
|
|
chr8_-_41522724
|
1.416
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr4_-_46391771
|
1.415
|
NM_000807
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr19_-_47164289
|
1.414
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr16_+_23847496
|
1.405
|
|
PRKCB
|
protein kinase C, beta
|
|
chr5_-_132948222
|
1.403
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr14_+_24540742
|
1.403
|
NM_006032
|
CPNE6
|
copine VI (neuronal)
|
|
chr1_-_26232890
|
1.391
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr16_+_58497628
|
1.383
|
|
NDRG4
|
NDRG family member 4
|
|
chr4_-_46391393
|
1.376
|
NM_001114175
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr5_+_87564729
|
1.374
|
|
LOC100505894
|
uncharacterized LOC100505894
|
|
chr15_-_44487468
|
1.360
|
|
FRMD5
|
FERM domain containing 5
|
|
chr1_+_233749749
|
1.357
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr11_-_119252364
|
1.357
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_26232643
|
1.353
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr13_-_102068705
|
1.347
|
NM_052867
|
NALCN
|
sodium leak channel, non-selective
|
|
chr1_-_40782826
|
1.340
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr20_-_61885802
|
1.322
|
NM_152864
|
NKAIN4
|
Na+/K+ transporting ATPase interacting 4
|
|
chr19_+_19322763
|
1.295
|
NM_004386
|
NCAN
|
neurocan
|
|
chr3_+_35721115
|
1.290
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr16_+_58497993
|
1.284
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr19_+_16771912
|
1.283
|
NM_024074
|
TMEM38A
|
transmembrane protein 38A
|
|
chr18_-_74844710
|
1.280
|
|
MBP
|
myelin basic protein
|
|
chr20_+_61273786
|
1.276
|
NM_016354
|
SLCO4A1
|
solute carrier organic anion transporter family, member 4A1
|
|
chr22_-_18923654
|
1.272
|
|
PRODH
|
proline dehydrogenase (oxidase) 1
|
|
chr5_+_87564840
|
1.251
|
|
LOC100505894
|
uncharacterized LOC100505894
|
|
chr11_-_790103
|
1.244
|
NM_016564
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr1_-_84464728
|
1.242
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr6_-_84419108
|
1.231
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr13_+_100633977
|
1.226
|
NM_007129
|
ZIC2
|
Zic family member 2
|
|
chr20_+_30407175
|
1.223
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr5_-_142000899
|
1.222
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr22_-_38840032
|
1.221
|
NM_004981
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr21_-_42219038
|
1.217
|
NM_001389
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr17_+_5972425
|
1.208
|
|
WSCD1
|
WSC domain containing 1
|
|
chr11_+_67183144
|
1.208
|
NM_001166222
NM_020811
|
CARNS1
|
carnosine synthase 1
|
|
chr19_+_18723666
|
1.207
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr1_-_9129666
|
1.206
|
NM_001135585
NM_003039
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr11_-_35441501
|
1.201
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr7_+_123249076
|
1.185
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr1_-_109849613
|
1.177
|
NM_001010985
|
MYBPHL
|
myosin binding protein H-like
|
|
chr21_-_42218587
|
1.177
|
|
DSCAM
|
Down syndrome cell adhesion molecule
|
|
chr21_-_39288661
|
1.177
|
NM_002240
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6
|
|
chr16_+_58497548
|
1.174
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr7_-_124405680
|
1.166
|
NM_005302
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr11_+_7273400
|
1.159
|
|
SYT9
|
synaptotagmin IX
|
|
chr8_-_91094924
|
1.158
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr12_+_56075418
|
1.158
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr16_+_333117
|
1.146
|
NM_006849
|
PDIA2
|
protein disulfide isomerase family A, member 2
|
|
chr11_+_6280903
|
1.145
|
NM_176875
|
CCKBR
|
cholecystokinin B receptor
|
|
chr7_-_82791799
|
1.142
|
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr17_+_7554415
|
1.142
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr2_+_73144603
|
1.140
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chr16_+_23847321
|
1.140
|
|
PRKCB
|
protein kinase C, beta
|
|
chr6_-_84419018
|
1.138
|
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr1_+_233749917
|
1.132
|
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr1_+_110693131
|
1.117
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr2_-_70781146
|
1.113
|
NM_001099691
NM_003236
|
TGFA
|
transforming growth factor, alpha
|
|
chr17_-_3599298
|
1.107
|
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr11_-_119234628
|
1.101
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_+_155146324
|
1.094
|
NM_025058
|
TRIM46
|
tripartite motif containing 46
|
|
chrX_+_37545008
|
1.093
|
NM_021083
|
XK
|
X-linked Kx blood group (McLeod syndrome)
|
|
chr1_-_145075636
|
1.093
|
NM_022359
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_+_125774271
|
1.092
|
NM_013264
|
DDX25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr1_+_57110745
|
1.086
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr17_-_29624249
|
1.083
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr4_-_46391461
|
1.081
|
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2
|
|
chr1_-_75139421
|
1.081
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr16_-_21289637
|
1.080
|
|
CRYM
|
crystallin, mu
|
|
chr15_-_38856830
|
1.079
|
NM_001128602
NM_005739
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated)
|
|
chr11_-_65816650
|
1.078
|
NM_033036
|
GAL3ST3
|
galactose-3-O-sulfotransferase 3
|
|
chr21_+_34398210
|
1.068
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr9_+_37650996
|
1.062
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr19_-_51472816
|
1.052
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr12_+_56075329
|
1.046
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr17_+_33474835
|
1.045
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr10_-_75410778
|
1.043
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr15_+_23810808
|
1.041
|
|
MKRN3
|
makorin ring finger protein 3
|
|
chr9_+_34990715
|
1.036
|
|
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5
|
|
chr7_-_51384458
|
1.030
|
NM_015198
|
COBL
|
cordon-bleu homolog (mouse)
|
|
chr2_-_209054772
|
1.026
|
NM_001099334
|
C2orf80
|
chromosome 2 open reading frame 80
|
|
chr2_-_239148626
|
1.021
|
NM_001142853
NM_018645
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr12_+_57943780
|
1.021
|
|
KIF5A
|
kinesin family member 5A
|
|
chr1_-_9129620
|
1.017
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr19_+_3933100
|
1.016
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr1_+_89990442
|
1.016
|
|
LRRC8B
|
leucine rich repeat containing 8 family, member B
|
|
chr14_+_100150596
|
1.009
|
NM_006668
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1
|
|
chr1_+_24742244
|
1.006
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr6_-_111136288
|
1.005
|
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr3_+_32148136
|
1.003
|
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr8_-_139509064
|
1.000
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr12_-_49392909
|
0.997
|
NM_015086
|
DDN
|
dendrin
|
|
chr20_+_44035203
|
0.996
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr12_+_57943806
|
0.994
|
|
KIF5A
|
kinesin family member 5A
|
|
chr20_+_44034632
|
0.992
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr10_-_73848530
|
0.988
|
NM_001244950
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr2_-_228029274
|
0.981
|
NM_000092
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr2_-_228028765
|
0.979
|
|
COL4A4
|
collagen, type IV, alpha 4
|
|
chr2_-_224467049
|
0.976
|
|
SCG2
|
secretogranin II
|
|
chr4_-_8160435
|
0.975
|
NM_001130083
NM_001130084
NM_001130085
NM_001130086
NM_001130087
NM_001130088
NM_032432
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr11_-_119252267
|
0.974
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr15_+_76352270
|
0.973
|
NM_152335
|
C15orf27
|
chromosome 15 open reading frame 27
|
|
chr7_-_82792127
|
0.972
|
NM_014510
NM_033026
|
PCLO
|
piccolo (presynaptic cytomatrix protein)
|
|
chr5_-_132948195
|
0.971
|
|
FSTL4
|
follistatin-like 4
|