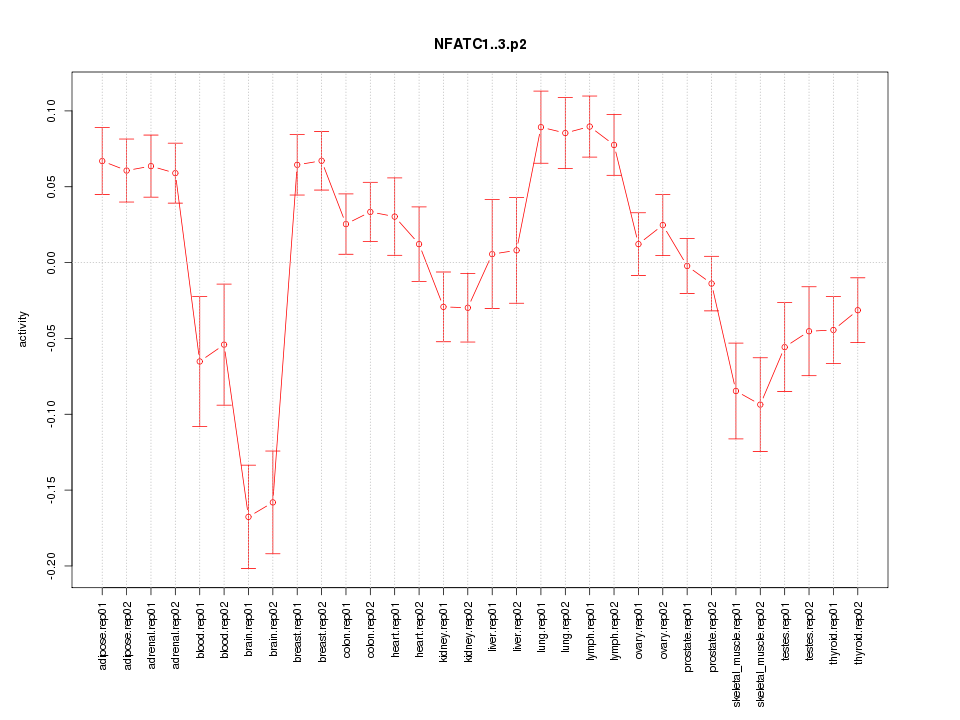
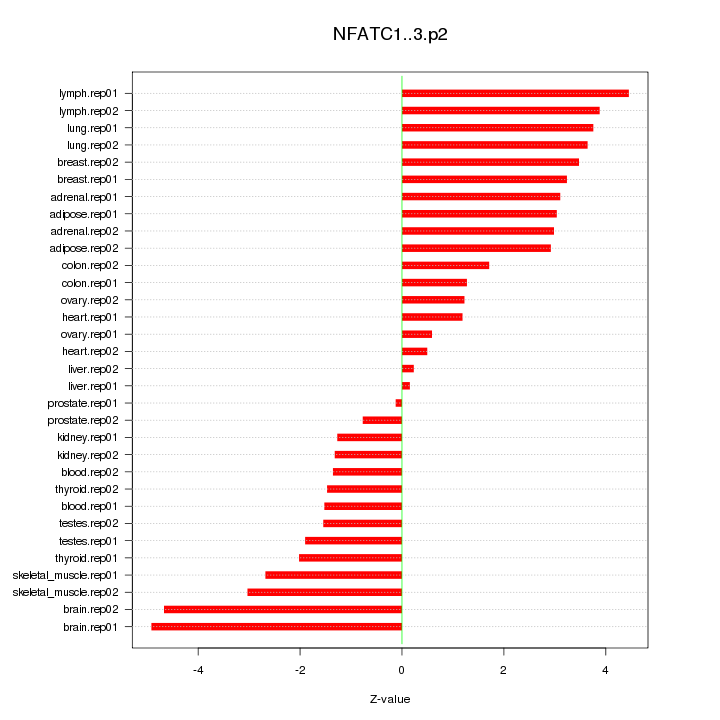
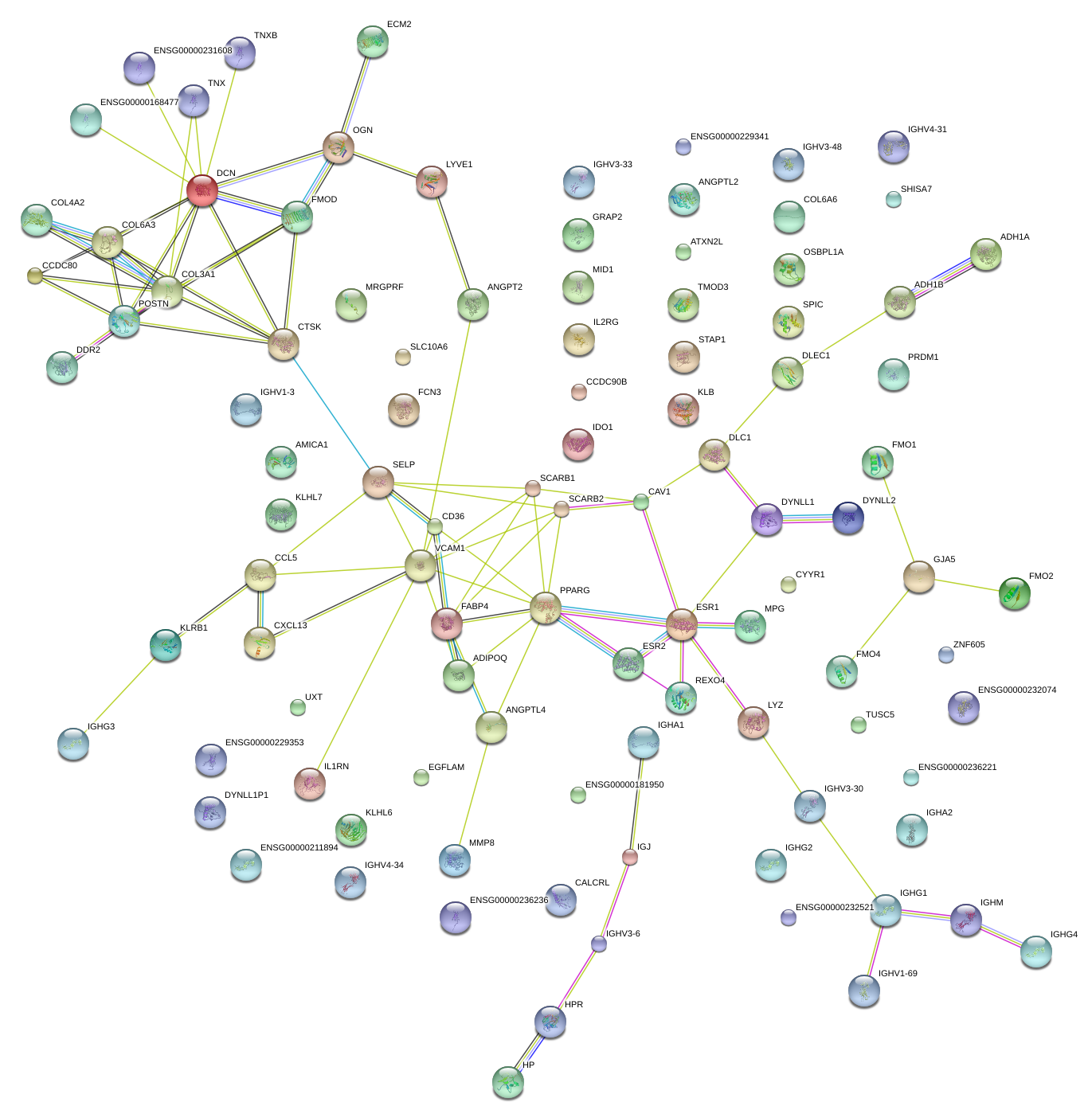
|
chr4_-_71532341
|
15.063
|
NM_144646
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr4_-_71532206
|
13.537
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr2_+_189839132
|
10.111
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr14_-_106573348
|
9.796
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr2_+_189839078
|
9.591
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr2_+_189839113
|
9.162
|
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr14_-_107122358
|
8.639
|
|
|
|
|
chr14_-_107218786
|
8.577
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr2_-_238322840
|
7.536
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322806
|
7.465
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_169599349
|
7.438
|
NM_003005
|
SELP
|
selectin P (granule membrane protein 140kDa, antigen CD62)
|
|
chr14_-_107013122
|
7.249
|
|
IGHV4-31
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr2_-_238322814
|
7.089
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_238322815
|
6.934
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr2_-_89165652
|
6.825
|
|
|
|
|
chr2_-_238322770
|
6.803
|
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr14_-_106994001
|
6.683
|
|
IGHV3-48
IGH@
IGHA1
IGHM
|
immunoglobulin heavy variable 3-48
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr3_-_112359962
|
6.481
|
NM_199511
NM_199512
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_171154387
|
6.299
|
NM_001460
|
FMO2
|
flavin containing monooxygenase 2 (non-functional)
|
|
chr3_-_112359863
|
6.054
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chrX_-_70331297
|
6.044
|
NM_000206
|
IL2RG
|
interleukin 2 receptor, gamma
|
|
chr1_-_203320185
|
5.451
|
|
FMOD
|
fibromodulin
|
|
chr17_+_48262816
|
5.328
|
|
|
|
|
chr14_-_106746056
|
5.283
|
|
|
|
|
chr21_-_27945267
|
5.211
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr8_-_82395401
|
5.197
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr4_-_100242487
|
5.174
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr1_-_203320287
|
5.074
|
NM_002023
|
FMOD
|
fibromodulin
|
|
chr1_-_203320249
|
5.023
|
|
|
|
|
chr14_-_106877689
|
4.986
|
|
IGHV4-31
IGH@
IGHA1
IGHA2
IGHG1
IGHG4
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 4 (G4m marker)
immunoglobulin heavy constant mu
|
|
chr14_-_107211131
|
4.720
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_-_91573264
|
4.660
|
NM_133503
|
DCN
|
decorin
|
|
chr2_-_188312872
|
4.617
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr14_-_106926393
|
4.471
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_+_80267782
|
4.456
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr14_-_107131217
|
4.433
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr3_+_186560478
|
4.410
|
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr7_+_80231503
|
4.254
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_-_102595623
|
4.228
|
NM_002424
|
MMP8
|
matrix metallopeptidase 8 (neutrophil collagenase)
|
|
chr1_+_162602198
|
4.131
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr17_+_1182956
|
4.090
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr6_+_152011630
|
4.081
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr2_+_113885145
|
4.070
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_+_186560462
|
4.051
|
NM_001177800
NM_004797
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr14_-_107170400
|
4.028
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_147232684
|
3.951
|
NM_181703
|
GJA5
|
gap junction protein, alpha 5, 40kDa
|
|
chr6_-_31981049
|
3.939
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr9_-_129884998
|
3.801
|
NM_012098
|
ANGPTL2
|
angiopoietin-like 2
|
|
chrX_-_10544852
|
3.706
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr11_-_10590078
|
3.705
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr4_-_100212085
|
3.642
|
NM_000667
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
|
|
chr1_-_150780705
|
3.584
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr5_+_38445577
|
3.582
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr14_-_107170076
|
3.502
|
|
IGHV1-69
IGHG1
|
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_+_69742140
|
3.468
|
|
LYZ
|
lysozyme
|
|
chr6_-_32013806
|
3.443
|
NM_032470
|
TNXB
|
tenascin XB
|
|
chr4_+_68424444
|
3.413
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr1_-_27701302
|
3.317
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chr3_-_183273407
|
3.276
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr8_-_13372183
|
3.275
|
NM_024767
NM_182643
|
DLC1
|
deleted in liver cancer 1
|
|
chr1_+_171217632
|
3.264
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr4_+_39408472
|
3.259
|
NM_175737
|
KLB
|
klotho beta
|
|
chr8_+_39771327
|
3.257
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr9_-_95298251
|
3.246
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr14_-_106539099
|
3.232
|
|
IGHA1
IGHG1
IGHG3
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr22_+_40297085
|
3.203
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr16_+_72088507
|
3.177
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr3_+_130279177
|
3.121
|
NM_001102608
|
COL6A6
|
collagen, type VI, alpha 6
|
|
chr13_+_111134933
|
3.121
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr15_+_52201590
|
3.087
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr2_+_113885137
|
3.067
|
NM_173842
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr17_-_34207308
|
3.051
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr3_+_12392993
|
3.051
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr11_-_118084073
|
3.046
|
NM_153206
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr7_+_116164838
|
3.020
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr1_+_101185195
|
3.019
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr6_+_106546736
|
3.010
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr9_-_95166827
|
3.009
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr4_+_78432905
|
3.008
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr7_+_80267922
|
2.997
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr13_-_38172862
|
2.978
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr8_-_6420454
|
2.976
|
NM_001118887
NM_001118888
NM_001147
|
ANGPT2
|
angiopoietin 2
|
|
chr7_+_80267972
|
2.944
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr11_-_68780667
|
2.904
|
NM_001098515
NM_145015
|
MRGPRF
|
MAS-related GPR, member F
|
|
chr14_-_106405633
|
2.872
|
|
IGHM
|
immunoglobulin heavy constant mu
|
|
chr4_-_87770267
|
2.859
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr7_+_23286384
|
2.831
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr9_+_12774988
|
2.783
|
NM_203403
|
C9orf150
|
chromosome 9 open reading frame 150
|
|
chr3_+_132316080
|
2.783
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr3_-_149051258
|
2.763
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr3_-_150920946
|
2.753
|
NM_013308
|
GPR171
|
G protein-coupled receptor 171
|
|
chr12_-_9760481
|
2.751
|
NM_002258
|
KLRB1
|
killer cell lectin-like receptor subfamily B, member 1
|
|
chr1_+_32739711
|
2.743
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr7_+_23286386
|
2.721
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_-_15038724
|
2.718
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr3_+_187943192
|
2.685
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_23286390
|
2.674
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr6_-_169654047
|
2.664
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr1_+_26644458
|
2.663
|
|
CD52
|
CD52 molecule
|
|
chr12_-_91505215
|
2.658
|
|
LUM
|
lumican
|
|
chr5_+_42565963
|
2.633
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr4_+_40198526
|
2.621
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr22_-_36236421
|
2.614
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr14_+_88471467
|
2.609
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr19_+_41281281
|
2.607
|
NM_006533
|
MIA
|
melanoma inhibitory activity
|
|
chr7_+_94024201
|
2.594
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr7_+_100199881
|
2.592
|
NM_002593
|
PCOLCE
|
procollagen C-endopeptidase enhancer
|
|
chr2_+_27301693
|
2.585
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr1_+_162602308
|
2.582
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr1_-_153518265
|
2.567
|
NM_002961
NM_019554
|
S100A4
|
S100 calcium binding protein A4
|
|
chr14_-_106552313
|
2.563
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr3_-_112358510
|
2.558
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr17_-_46667596
|
2.535
|
|
HOXB3
|
homeobox B3
|
|
chr14_-_106967080
|
2.529
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr4_+_102734982
|
2.492
|
NM_001083907
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1
|
|
chr8_-_86290333
|
2.491
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr12_-_9268475
|
2.476
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_+_86046434
|
2.473
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr12_+_13349716
|
2.466
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_-_9268513
|
2.465
|
|
A2M
|
alpha-2-macroglobulin
|
|
chrX_+_2670092
|
2.460
|
NM_001141919
NM_001141920
NM_175569
|
XGPY2
XG
|
Xg pseudogene, Y-linked 2
Xg blood group
|
|
chr1_+_32712817
|
2.460
|
NM_032648
|
FAM167B
|
family with sequence similarity 167, member B
|
|
chr3_-_112358758
|
2.458
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr7_-_27166638
|
2.455
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr7_-_131241321
|
2.454
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr5_+_54398473
|
2.451
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr12_+_69742130
|
2.443
|
NM_000239
|
LYZ
|
lysozyme
|
|
chr22_+_33196801
|
2.414
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr19_+_3178735
|
2.400
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|
|
chr4_-_76928601
|
2.386
|
NM_002416
|
CXCL9
|
chemokine (C-X-C motif) ligand 9
|
|
chr3_-_112359277
|
2.378
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_26644380
|
2.366
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr20_+_12989616
|
2.353
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr7_+_23286401
|
2.342
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr3_+_14860468
|
2.342
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr3_-_151176496
|
2.330
|
NM_178822
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr5_-_158526698
|
2.329
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr21_-_28339405
|
2.302
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr1_+_117297056
|
2.278
|
NM_001767
|
CD2
|
CD2 molecule
|
|
chr7_-_15726026
|
2.242
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr14_-_106691698
|
2.239
|
|
|
|
|
chr14_+_63671101
|
2.236
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr8_-_95274540
|
2.235
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_-_112359251
|
2.234
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_-_85462622
|
2.230
|
NM_153259
|
MCOLN2
|
mucolipin 2
|
|
chr12_+_12938540
|
2.230
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr12_+_13349601
|
2.228
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr10_+_35484829
|
2.222
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr13_-_52980628
|
2.222
|
NM_018676
NM_199263
|
THSD1
|
thrombospondin, type I, domain containing 1
|
|
chr13_+_102104941
|
2.209
|
NM_004791
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains)
|
|
chr12_+_13349720
|
2.191
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr22_-_19512859
|
2.172
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr1_-_157522151
|
2.164
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr15_+_73976689
|
2.157
|
|
CD276
|
CD276 molecule
|
|
chrX_+_15518899
|
2.124
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr14_-_106967528
|
2.124
|
|
IGHA1
IGHA2
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_171621750
|
2.121
|
NM_000261
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response
|
|
chr6_-_112575766
|
2.110
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr5_-_172755418
|
2.108
|
|
STC2
|
stanniocalcin 2
|
|
chr14_-_106641585
|
2.104
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr15_+_81293286
|
2.093
|
NM_022566
|
MESDC1
|
mesoderm development candidate 1
|
|
chr10_+_17851342
|
2.088
|
NM_002438
|
MRC1
|
mannose receptor, C type 1
|
|
chr17_-_42908178
|
2.086
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr12_-_102872329
|
2.082
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr6_-_46825942
|
2.078
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr6_-_112575708
|
2.078
|
|
LAMA4
|
laminin, alpha 4
|
|
chr17_-_34344933
|
2.071
|
NM_005064
NM_145898
|
CCL23
|
chemokine (C-C motif) ligand 23
|
|
chr1_-_54483802
|
2.066
|
NM_001010978
|
LDLRAD1
|
low density lipoprotein receptor class A domain containing 1
|
|
chr8_-_95274520
|
2.063
|
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr3_+_45984972
|
2.061
|
NM_006564
|
CXCR6
|
chemokine (C-X-C motif) receptor 6
|
|
chr12_+_86268072
|
2.046
|
NM_006183
|
NTS
|
neurotensin
|
|
chr12_+_57388229
|
2.036
|
NM_007264
|
GPR182
|
G protein-coupled receptor 182
|
|
chr14_-_107048697
|
2.035
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr12_-_91505541
|
2.035
|
NM_002345
|
LUM
|
lumican
|
|
chr12_-_52886971
|
2.029
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr20_+_24929904
|
2.024
|
|
CST7
|
cystatin F (leukocystatin)
|
|
chr6_-_138189313
|
2.023
|
|
|
|
|
chr3_+_361365
|
2.020
|
NM_001253388
|
CHL1
|
cell adhesion molecule with homology to L1CAM (close homolog of L1)
|
|
chr2_-_216259197
|
1.994
|
|
FN1
|
fibronectin 1
|
|
chr3_-_112359565
|
1.986
|
|
CCDC80
|
coiled-coil domain containing 80
|
|
chr1_+_45274153
|
1.983
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr14_-_24977421
|
1.972
|
NM_001836
|
CMA1
|
chymase 1, mast cell
|
|
chr10_+_81370694
|
1.972
|
NM_001093770
NM_001164644
NM_001164645
NM_001164646
NM_001164647
NM_005411
|
SFTPA1
|
surfactant protein A1
|
|
chr6_-_169653922
|
1.945
|
|
THBS2
|
thrombospondin 2
|
|
chr14_-_106791054
|
1.944
|
|
IGHV4-31
IGHV3-30
IGHA1
IGHD
IGHG1
IGHG2
IGHG3
IGHM
SCFV
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-30
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 2 (G2m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
single-chain Fv fragment
|
|
chr1_+_159175048
|
1.941
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr17_-_38520941
|
1.918
|
NM_152219
|
GJD3
|
gap junction protein, delta 3, 31.9kDa
|
|
chr10_-_81320141
|
1.917
|
NM_001098668
|
SFTPA1
SFTPA2
|
surfactant protein A1
surfactant protein A2
|
|
chr11_-_118095708
|
1.917
|
NM_001098526
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr4_+_124317884
|
1.916
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr14_-_64749530
|
1.913
|
NM_001214902
NM_001214903
|
ESR2
|
estrogen receptor 2 (ER beta)
|
|
chr3_-_149095517
|
1.910
|
NM_014220
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr7_-_16844606
|
1.906
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr4_+_90816002
|
1.900
|
NM_007351
|
MMRN1
|
multimerin 1
|
|
chr6_-_46922533
|
1.900
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr3_+_122334461
|
1.899
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr15_+_74466024
|
1.899
|
NM_005545
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr4_-_152147520
|
1.892
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr4_+_74606222
|
1.881
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr15_+_73976621
|
1.872
|
NM_001024736
NM_025240
|
CD276
|
CD276 molecule
|