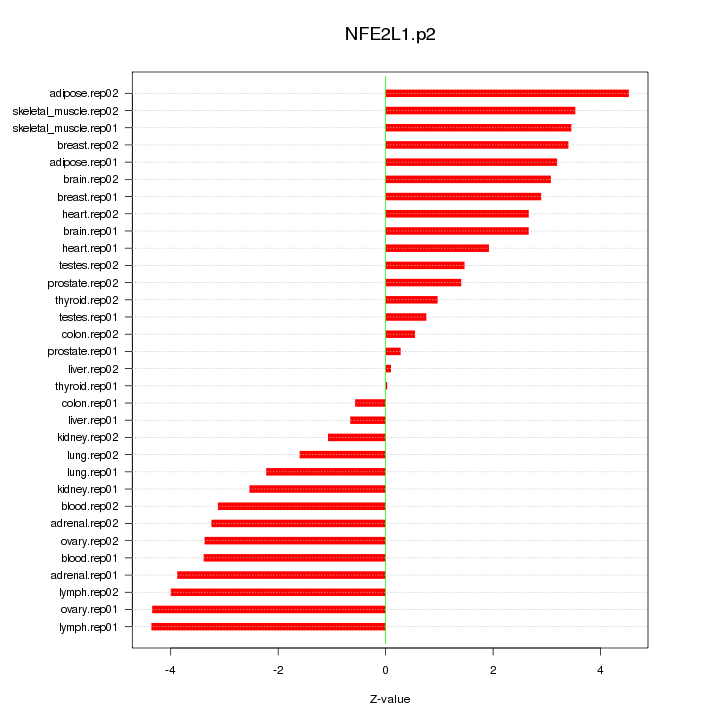
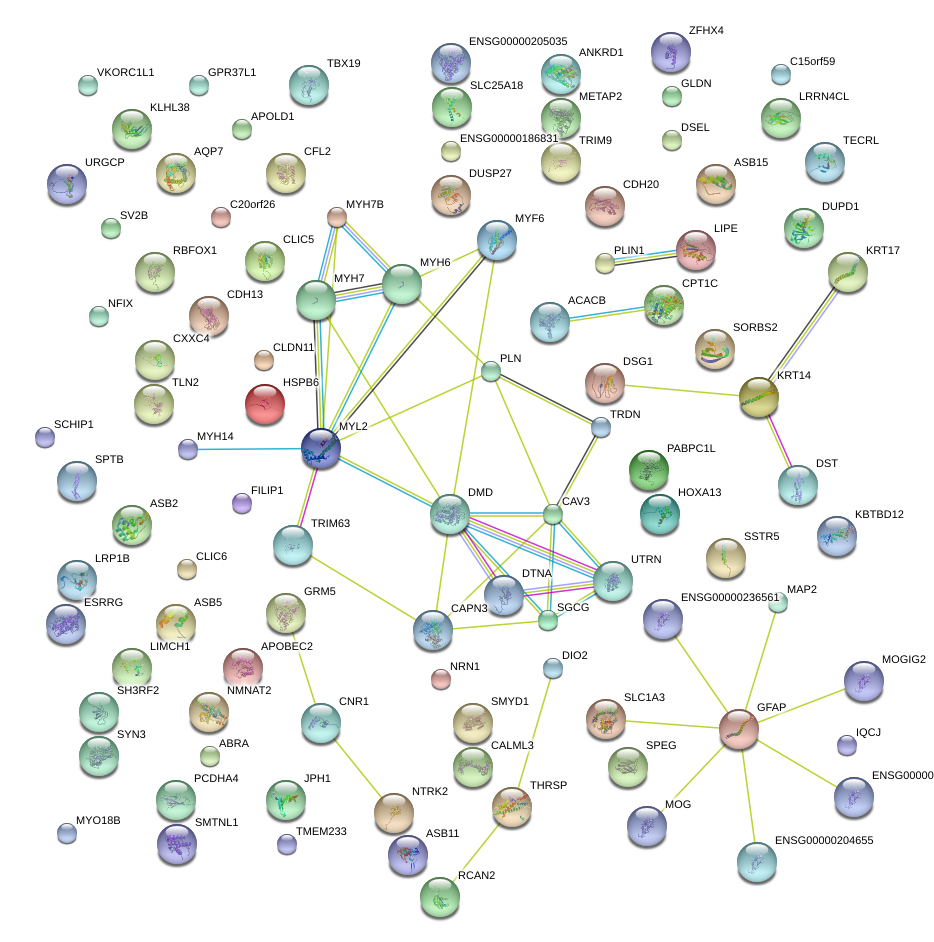
|
chr2_+_88367298
|
8.954
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr5_+_36608430
|
6.513
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_+_12938540
|
6.437
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr6_-_56716412
|
5.470
|
|
DST
|
dystonin
|
|
chr7_+_123249076
|
5.392
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr12_+_81101407
|
4.972
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr18_+_59157774
|
4.858
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr15_+_91769329
|
4.825
|
|
SV2B
|
synaptic vesicle glycoprotein 2B
|
|
chr3_+_127641901
|
4.596
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr1_-_26393984
|
4.227
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chrX_-_32430370
|
4.123
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr17_-_42992852
|
4.048
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chr12_+_109577201
|
3.999
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr19_+_50706868
|
3.850
|
NM_001077186
NM_001145809
NM_024729
|
MYH14
|
myosin, heavy chain 14, non-muscle
|
|
chr6_-_46047978
|
3.848
|
NM_001114086
|
CLIC5
|
chloride intracellular channel 5
|
|
chr14_-_65289812
|
3.772
|
NM_000347
NM_001024858
|
SPTB
|
spectrin, beta, erythrocytic
|
|
chr4_-_177190258
|
3.708
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr6_+_41020939
|
3.659
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chr3_+_158991035
|
3.657
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr2_-_142889269
|
3.654
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr17_+_17398334
|
3.610
|
|
|
|
|
chr4_-_65275000
|
3.493
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr19_-_36019194
|
3.380
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr15_+_62939509
|
3.375
|
NM_015059
|
TLN2
|
talin 2
|
|
chr18_+_32398225
|
3.346
|
NM_001198942
NM_001198944
NM_032980
NM_032981
|
DTNA
|
dystrobrevin, alpha
|
|
chr6_-_56716713
|
3.281
|
|
DST
|
dystonin
|
|
chr20_+_20037289
|
3.266
|
NM_001167816
|
C20orf26
|
chromosome 20 open reading frame 26
|
|
chr11_+_77774902
|
3.233
|
NM_003251
|
THRSP
|
thyroid hormone responsive
|
|
chr1_-_217250230
|
3.154
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_118869441
|
3.151
|
NM_002667
|
PLN
|
phospholamban
|
|
chr4_-_105412466
|
3.150
|
NM_025212
|
CXXC4
|
CXXC finger protein 4
|
|
chr14_-_35182944
|
3.149
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr8_-_124665165
|
3.084
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr15_+_51633712
|
3.057
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr11_+_57310113
|
3.026
|
NM_001105565
|
SMTNL1
|
smoothelin-like 1
|
|
chr16_+_82660390
|
3.016
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr11_-_62457001
|
2.989
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr19_-_36247917
|
2.987
|
NM_144617
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr8_-_107782417
|
2.984
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr3_+_8775485
|
2.968
|
NM_001234
NM_033337
|
CAV3
|
caveolin 3
|
|
chr4_-_186577858
|
2.949
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_123957941
|
2.941
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr1_+_202091971
|
2.934
|
NM_004767
|
GPR37L1
|
G protein-coupled receptor 37 like 1
|
|
chr14_-_80677839
|
2.930
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chrX_-_15333726
|
2.921
|
NM_001201583
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr4_-_186578122
|
2.893
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_46293404
|
2.885
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr22_+_26138110
|
2.880
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr13_+_23755059
|
2.863
|
NM_000231
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein)
|
|
chr14_-_51561657
|
2.830
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr16_+_7382750
|
2.825
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chrX_-_15332680
|
2.812
|
NM_001012428
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr3_+_158991090
|
2.793
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr19_+_13135745
|
2.792
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr6_-_88854989
|
2.738
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr18_+_28898051
|
2.735
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr7_-_27239724
|
2.728
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr10_-_92680784
|
2.723
|
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr5_+_145317297
|
2.717
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr16_+_1128780
|
2.700
|
NM_001053
|
SSTR5
|
somatostatin receptor 5
|
|
chr22_-_33402648
|
2.643
|
NM_003490
NM_133633
|
SYN3
|
synapsin III
|
|
chr11_-_88781039
|
2.640
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr17_-_39780762
|
2.626
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr14_-_23904849
|
2.608
|
NM_000257
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta
|
|
chr19_-_42931500
|
2.601
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr6_-_6007097
|
2.552
|
|
NRN1
|
neuritin 1
|
|
chr2_+_220309373
|
2.526
|
|
SPEG
|
SPEG complex locus
|
|
chr3_+_170136652
|
2.501
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr14_-_94423766
|
2.496
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr10_+_5566923
|
2.487
|
NM_005185
|
CALML3
|
calmodulin-like 3
|
|
chr10_-_92681025
|
2.478
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr8_+_77765817
|
2.475
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr17_-_39743085
|
2.455
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr18_-_65182507
|
2.450
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr9_+_87283453
|
2.443
|
NM_001018064
NM_006180
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr22_+_18043147
|
2.434
|
NM_031481
|
SLC25A18
|
solute carrier family 25 (mitochondrial carrier), member 18
|
|
chr12_+_120031263
|
2.429
|
NM_001136534
|
TMEM233
|
transmembrane protein 233
|
|
chr1_-_183387501
|
2.429
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr15_+_42651697
|
2.419
|
NM_000070
NM_024344
NM_173087
|
CAPN3
|
calpain 3, (p94)
|
|
chr15_-_74043815
|
2.419
|
NM_001039614
|
C15orf59
|
chromosome 15 open reading frame 59
|
|
chr9_-_33395265
|
2.417
|
|
AQP7
|
aquaporin 7
|
|
chrX_-_33357725
|
2.409
|
NM_000109
|
DMD
|
dystrophin
|
|
chr7_+_65419441
|
2.374
|
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1
|
|
chr1_+_167064086
|
2.342
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr6_+_29624757
|
2.316
|
NM_001008228
NM_001008229
NM_001170418
NM_002433
NM_206809
NM_206810
NM_206811
NM_206812
NM_206814
|
MOG
|
myelin oligodendrocyte glycoprotein
|
|
chr12_-_111358353
|
2.290
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr16_+_82660573
|
2.286
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr6_-_76203256
|
2.194
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr4_+_41700736
|
2.193
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_+_210444364
|
2.193
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr15_-_90222572
|
2.186
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr8_-_75233461
|
2.178
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr17_-_39768939
|
2.145
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr5_+_53751430
|
2.132
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr3_-_69171536
|
2.128
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr11_+_12695851
|
2.126
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr17_+_53344966
|
2.107
|
|
HLF
|
hepatic leukemia factor
|
|
chr6_-_46825942
|
2.104
|
|
GPR116
|
G protein-coupled receptor 116
|
|
chr10_-_76868853
|
2.104
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr7_-_113559048
|
2.102
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chr2_-_211168315
|
2.095
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr3_+_29322802
|
2.088
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr6_-_139613114
|
2.061
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr2_-_2334887
|
2.059
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr2_+_170366211
|
2.056
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr18_+_32073245
|
2.051
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr5_-_150727088
|
2.046
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr6_+_117586720
|
2.039
|
NM_153453
NM_182645
|
VGLL2
|
vestigial like 2 (Drosophila)
|
|
chr9_-_14313509
|
2.037
|
|
NFIB
|
nuclear factor I/B
|
|
chrX_+_105969875
|
2.037
|
NM_194463
|
RNF128
|
ring finger protein 128
|
|
chr5_-_138210992
|
2.025
|
|
CTNNA1
LRRTM2
|
catenin (cadherin-associated protein), alpha 1, 102kDa
leucine rich repeat transmembrane neuronal 2
|
|
chr17_+_33474835
|
2.023
|
NM_001033576
NM_173167
|
UNC45B
|
unc-45 homolog B (C. elegans)
|
|
chr18_-_70532933
|
2.019
|
NM_138999
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr1_-_168698412
|
2.012
|
NM_001937
|
DPT
|
dermatopontin
|
|
chr6_-_46293628
|
2.007
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chr6_+_72922513
|
2.007
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr16_+_82660641
|
1.998
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr3_-_58563070
|
1.993
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr6_-_6007632
|
1.987
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr12_+_13349716
|
1.987
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr12_-_71148063
|
1.984
|
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_+_71403040
|
1.977
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr11_+_17757494
|
1.968
|
NM_001112741
NM_004976
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1
|
|
chr5_+_161277702
|
1.949
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr7_+_136553398
|
1.942
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr2_-_2335080
|
1.935
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr7_-_27224762
|
1.922
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chr12_+_112856701
|
1.919
|
|
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr3_-_151987332
|
1.913
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr14_+_74083547
|
1.904
|
NM_001037162
|
ACOT6
|
acyl-CoA thioesterase 6
|
|
chr5_+_6448735
|
1.902
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr5_+_6449114
|
1.902
|
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr3_-_179754516
|
1.894
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr4_-_111563278
|
1.880
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chrX_+_18709044
|
1.872
|
NM_006240
NM_152224
NM_152226
|
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1
|
|
chr2_+_172950207
|
1.856
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr15_-_83349134
|
1.855
|
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr11_-_62689011
|
1.852
|
NM_000738
|
CHRM1
|
cholinergic receptor, muscarinic 1
|
|
chr16_-_66959418
|
1.846
|
NM_001128850
NM_004165
|
RRAD
|
Ras-related associated with diabetes
|
|
chr19_+_13135809
|
1.842
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr9_+_35042222
|
1.842
|
NM_001040410
NM_203299
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr17_+_1182956
|
1.840
|
NM_172367
|
TUSC5
|
tumor suppressor candidate 5
|
|
chr3_-_192126837
|
1.839
|
NM_021032
|
FGF12
|
fibroblast growth factor 12
|
|
chr6_+_17281773
|
1.814
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr18_+_29171729
|
1.808
|
NM_000371
|
TTR
|
transthyretin
|
|
chr14_-_21270537
|
1.795
|
NM_198232
|
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chr22_-_36013303
|
1.793
|
NM_005368
NM_203378
|
MB
|
myoglobin
|
|
chr12_+_13349720
|
1.786
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr7_-_8302141
|
1.784
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr10_+_88428263
|
1.781
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr11_+_124120422
|
1.778
|
NM_001002905
|
OR8G1
|
olfactory receptor, family 8, subfamily G, member 1
|
|
chr6_-_46293215
|
1.777
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr12_-_57634474
|
1.773
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr2_-_211179765
|
1.765
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chrX_+_70443055
|
1.762
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chrX_+_99899162
|
1.756
|
NM_014467
|
SRPX2
|
sushi-repeat containing protein, X-linked 2
|
|
chr13_-_33760215
|
1.755
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr17_-_38256548
|
1.751
|
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr17_-_62050200
|
1.751
|
NM_000334
|
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit
|
|
chr19_-_49658645
|
1.749
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr4_-_185726865
|
1.748
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr6_-_112575766
|
1.743
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr1_+_84767288
|
1.731
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_+_10857867
|
1.723
|
NM_014229
|
SLC6A11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr9_-_129870311
|
1.714
|
|
ANGPTL2
|
angiopoietin-like 2
|
|
chrX_-_31284946
|
1.714
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr16_+_15596122
|
1.710
|
NM_001142469
|
C16orf45
|
chromosome 16 open reading frame 45
|
|
chr3_+_35722428
|
1.710
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr4_-_47840058
|
1.708
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr1_-_144994908
|
1.702
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr12_-_57642922
|
1.700
|
|
|
|
|
chr7_+_27225009
|
1.698
|
|
HOXA11-AS1
|
HOXA11 antisense RNA 1 (non-protein coding)
|
|
chr8_-_72268720
|
1.696
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_+_82173574
|
1.692
|
NM_001243778
NM_001243782
|
C10orf58
|
chromosome 10 open reading frame 58
|
|
chr3_-_12200339
|
1.672
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr15_-_71184611
|
1.672
|
NM_020147
|
THAP10
|
THAP domain containing 10
|
|
chr12_-_49392909
|
1.671
|
NM_015086
|
DDN
|
dendrin
|
|
chr6_+_17393610
|
1.662
|
NM_006366
|
CAP2
|
CAP, adenylate cyclase-associated protein, 2 (yeast)
|
|
chr19_-_15083729
|
1.654
|
NM_005071
|
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6
|
|
chr7_-_135433366
|
1.643
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chrX_-_21776158
|
1.640
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr12_+_13349601
|
1.636
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr22_-_32651317
|
1.627
|
NM_014227
|
SLC5A4
|
solute carrier family 5 (low affinity glucose cotransporter), member 4
|
|
chr2_-_193059249
|
1.619
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr9_-_140115774
|
1.611
|
NM_031297
|
RNF208
|
ring finger protein 208
|
|
chr4_-_187527219
|
1.611
|
|
|
|
|
chr11_+_26353677
|
1.602
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr20_-_48099178
|
1.598
|
NM_004975
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1
|
|
chr5_-_95748688
|
1.596
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr1_-_159505796
|
1.595
|
NM_001004469
|
OR10J5
|
olfactory receptor, family 10, subfamily J, member 5
|
|
chr1_-_59248287
|
1.591
|
|
JUN
|
jun proto-oncogene
|
|
chrX_+_28605558
|
1.588
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr22_+_38597938
|
1.584
|
NM_001161572
NM_001161574
NM_012323
|
MAFF
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian)
|
|
chr11_-_47207425
|
1.578
|
NM_001184975
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr8_-_33457491
|
1.572
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr3_+_119316694
|
1.570
|
NM_001206960
NM_001206961
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr12_+_18891040
|
1.569
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr4_-_186456580
|
1.565
|
NM_001114107
NM_014476
|
PDLIM3
|
PDZ and LIM domain 3
|
|
chr5_+_175308843
|
1.560
|
|
CPLX2
|
complexin 2
|
|
chr11_-_19223516
|
1.558
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|