|
chr4_+_74269971
|
22.835
|
NM_000477
|
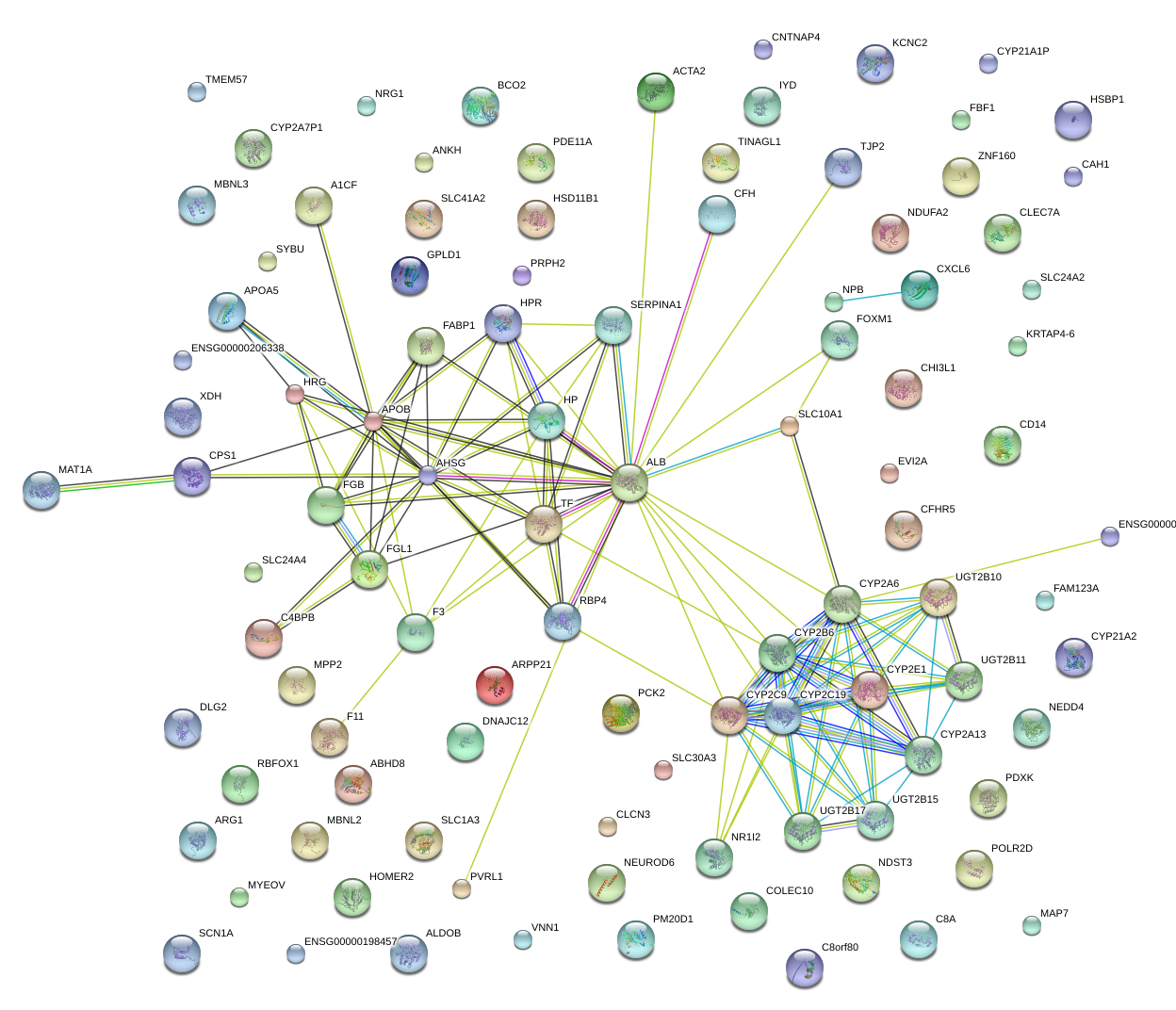
ALB
|
albumin
|
|
chr10_+_135340865
|
20.788
|
NM_000773
|
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1
|
|
chr10_-_95360959
|
19.989
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr3_+_133494275
|
15.858
|
|
TF
|
transferrin
|
|
chr10_-_82049178
|
12.715
|
NM_000429
|
MAT1A
|
methionine adenosyltransferase I, alpha
|
|
chr3_+_186383770
|
11.244
|
NM_000412
|
HRG
|
histidine-rich glycoprotein
|
|
chr1_+_57320442
|
11.132
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr4_+_155484131
|
10.480
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr1_+_196946666
|
8.882
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr9_-_104198045
|
8.699
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr14_-_70263871
|
7.861
|
NM_003049
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1
|
|
chr19_+_41497182
|
7.837
|
NM_000767
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr8_-_17752847
|
7.823
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr19_+_41430123
|
7.127
|
|
CYP2B7P1
|
cytochrome P450, family 2, subfamily B, polypeptide 7 pseudogene 1
|
|
chr1_-_203155910
|
6.875
|
NM_001276
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr1_-_203155813
|
6.553
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr1_-_203155745
|
6.520
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr1_-_203155827
|
6.410
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr19_-_41388656
|
6.375
|
NM_000764
NM_030589
|
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7
|
|
chr6_+_131894343
|
6.250
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr2_-_31637598
|
6.059
|
NM_000379
|
XDH
|
xanthine dehydrogenase
|
|
chr1_-_203155867
|
5.990
|
|
CHI3L1
|
chitinase 3-like 1 (cartilage glycoprotein-39)
|
|
chr3_+_119501556
|
5.930
|
NM_022002
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr19_+_41497284
|
5.460
|
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6
|
|
chr4_+_187187342
|
5.140
|
|
F11
|
coagulation factor XI
|
|
chr2_-_31637553
|
5.111
|
|
XDH
|
xanthine dehydrogenase
|
|
chr11_-_116663106
|
5.077
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr3_+_186330844
|
5.076
|
NM_001622
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr8_-_110661007
|
4.782
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr7_-_31380507
|
4.766
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr6_-_133035180
|
4.534
|
NM_004666
|
VNN1
|
vanin 1
|
|
chr5_-_140013285
|
4.441
|
NM_001040021
NM_001174104
NM_001174105
|
CD14
|
CD14 molecule
|
|
chr10_-_52645430
|
4.277
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr1_+_207262211
|
4.251
|
NM_001017365
NM_001017366
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr19_+_41594367
|
4.155
|
NM_000766
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13
|
|
chr1_+_25757381
|
4.074
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr4_+_118955499
|
4.043
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr14_+_24563369
|
3.938
|
NM_001018073
NM_004563
|
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr6_-_24489738
|
3.830
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr12_-_75603510
|
3.676
|
NM_139136
NM_139137
NM_153748
|
KCNC2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2
|
|
chr16_+_76311175
|
3.634
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr4_-_69536314
|
3.611
|
NM_001076
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15
|
|
chr5_-_140012730
|
3.538
|
|
CD14
|
CD14 molecule
|
|
chr6_-_136847609
|
3.486
|
|
MAP7
|
microtubule-associated protein 7
|
|
chr12_-_105322056
|
3.343
|
NM_032148
|
SLC41A2
|
solute carrier family 41, member 2
|
|
chr6_-_136847078
|
3.319
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr3_+_186330859
|
3.305
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr8_-_27941387
|
3.042
|
NM_001010906
|
C8orf80
|
chromosome 8 open reading frame 80
|
|
chr11_+_69061599
|
2.804
|
NM_138768
|
MYEOV
|
myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas)
|
|
chr16_+_72097124
|
2.736
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr2_+_211421322
|
2.723
|
NM_001875
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr13_+_97999289
|
2.694
|
|
|
|
|
chr2_-_21228189
|
2.668
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr10_-_69597768
|
2.622
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr5_-_14871555
|
2.619
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr1_+_209878135
|
2.611
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr2_-_167005641
|
2.582
|
NM_001202435
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr1_+_196621007
|
2.581
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr10_+_96522457
|
2.487
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr15_-_83621350
|
2.451
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr5_-_140013034
|
2.436
|
NM_000591
|
CD14
|
CD14 molecule
|
|
chr4_+_170581212
|
2.408
|
NM_001243374
|
CLCN3
|
chloride channel 3
|
|
chr19_-_17414083
|
2.345
|
NM_024527
|
ABHD8
|
abhydrolase domain containing 8
|
|
chr21_+_45138973
|
2.291
|
NM_003681
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr14_-_94849552
|
2.275
|
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr11_-_119599253
|
2.255
|
NM_002855
NM_203285
NM_203286
|
PVRL1
|
poliovirus receptor-related 1 (herpesvirus entry mediator C)
|
|
chr9_+_71819926
|
2.209
|
NM_001170415
NM_001170416
|
TJP2
|
tight junction protein 2 (zona occludens 2)
|
|
chr12_-_10282680
|
2.206
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr6_+_150690027
|
2.201
|
NM_001164694
NM_001164695
NM_203395
|
IYD
|
iodotyrosine deiodinase
|
|
chrX_-_131547536
|
2.169
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr13_-_25746304
|
2.102
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chr8_+_120079423
|
2.038
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr2_-_88427535
|
2.037
|
NM_001443
|
FABP1
|
fatty acid binding protein 1, liver
|
|
chr11_+_112047088
|
1.960
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr5_+_36606667
|
1.914
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_-_83984230
|
1.913
|
NM_001142700
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr3_+_186330711
|
1.859
|
|
AHSG
|
alpha-2-HS-glycoprotein
|
|
chr13_+_97874540
|
1.858
|
NM_144778
NM_207304
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr9_-_19786886
|
1.857
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr2_-_178753464
|
1.853
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chr4_-_110354914
|
1.844
|
|
1/2-SBSRNA4
|
lncRNA_BC009800
|
|
chr2_-_105467912
|
1.843
|
|
LOC100506421
|
uncharacterized LOC100506421
|
|
chr3_+_151488214
|
1.835
|
|
LOC201651
|
arylacetamide deacetylase (esterase) pseudogene
|
|
chr16_+_7382750
|
1.830
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_-_29648631
|
1.796
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr15_-_56209305
|
1.794
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr4_+_74702272
|
1.792
|
NM_002993
|
CXCL6
|
chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2)
|
|
chr3_-_190580464
|
1.763
|
NM_001146686
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr1_-_161207879
|
1.735
|
NM_001077469
NM_001077470
NM_001077471
NM_001077472
NM_001077473
NM_001077474
NM_001077475
NM_001077476
NM_001077477
NM_001077478
NM_001077479
NM_001077480
NM_001077481
NM_001077482
NM_005122
|
NR1I3
|
nuclear receptor subfamily 1, group I, member 3
|
|
chr4_-_176733901
|
1.705
|
|
GPM6A
|
glycoprotein M6A
|
|
chr7_-_111428944
|
1.669
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr2_+_201756639
|
1.669
|
NM_001142356
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae)
|
|
chr3_-_58200397
|
1.637
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr5_-_87969135
|
1.627
|
|
LINC00461
|
long intergenic non-protein coding RNA 461
|
|
chr6_+_134758840
|
1.626
|
|
LOC154092
|
uncharacterized LOC154092
|
|
chr21_+_45138987
|
1.624
|
|
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr13_+_97928414
|
1.588
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr16_+_72088507
|
1.582
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr5_+_36606456
|
1.579
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr18_-_48346297
|
1.564
|
NM_001127175
NM_001127176
NM_031939
|
MRO
|
maestro
|
|
chr12_-_7656285
|
1.538
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr11_+_123431036
|
1.534
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr8_+_99076749
|
1.532
|
NM_001170806
NM_173549
|
C8orf47
|
chromosome 8 open reading frame 47
|
|
chr3_+_101546833
|
1.474
|
NM_001005474
|
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta
|
|
chr16_-_31439650
|
1.473
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr20_+_58296264
|
1.471
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr13_-_101185944
|
1.467
|
NM_033110
|
A2LD1
|
AIG2-like domain 1
|
|
chr7_+_142982039
|
1.457
|
NM_001242773
NM_001242774
NM_001242775
NM_001242776
NM_001242777
NM_153345
|
TMEM139
|
transmembrane protein 139
|
|
chr10_+_5238756
|
1.449
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr2_+_102927961
|
1.435
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr8_-_110620220
|
1.417
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_156696351
|
1.328
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr16_-_55866972
|
1.326
|
|
CES1
|
carboxylesterase 1
|
|
chr17_+_61678218
|
1.324
|
NM_016360
|
TACO1
|
translational activator of mitochondrially encoded cytochrome c oxidase I
|
|
chr17_-_38821286
|
1.310
|
NM_152349
|
KRT222
|
keratin 222
|
|
chr3_-_169487602
|
1.295
|
NM_032487
|
ARPM1
|
actin related protein M1
|
|
chr2_+_179184970
|
1.292
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr8_+_24774587
|
1.278
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chrX_+_38663450
|
1.278
|
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr8_-_72274278
|
1.266
|
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr3_+_128532267
|
1.242
|
|
|
|
|
chr17_-_61850953
|
1.219
|
NM_020198
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr3_-_194188967
|
1.211
|
NM_024524
|
ATP13A3
|
ATPase type 13A3
|
|
chr1_-_110155672
|
1.190
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr5_-_43557520
|
1.180
|
NM_183323
|
PAIP1
|
poly(A) binding protein interacting protein 1
|
|
chrX_+_118108575
|
1.175
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr4_-_70518964
|
1.145
|
NM_001252274
NM_001252275
NM_006798
|
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus
|
|
chr1_-_112106562
|
1.136
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr17_-_61850890
|
1.133
|
|
CCDC47
|
coiled-coil domain containing 47
|
|
chr15_-_90222572
|
1.129
|
NM_001145311
NM_002666
|
PLIN1
|
perilipin 1
|
|
chr18_-_24722757
|
1.125
|
NM_001243848
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr19_+_3762650
|
1.122
|
NM_172251
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr10_-_74283693
|
1.110
|
NM_001195519
|
MICU1
|
mitochondrial calcium uptake 1
|
|
chrX_+_38663072
|
1.103
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr4_-_70080448
|
1.089
|
NM_001073
|
UGT2B11
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B11
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr1_-_54411245
|
1.076
|
NM_016126
|
HSPB11
|
heat shock protein family B (small), member 11
|
|
chr10_+_5005601
|
1.065
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr2_+_201473942
|
1.045
|
|
AOX1
|
aldehyde oxidase 1
|
|
chr22_-_50523853
|
1.023
|
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1
|
|
chr8_+_67405490
|
1.021
|
NM_152765
|
C8orf46
|
chromosome 8 open reading frame 46
|
|
chr1_+_232940637
|
1.019
|
NM_019090
|
KIAA1383
|
KIAA1383
|
|
chr19_+_3762671
|
1.019
|
|
MRPL54
|
mitochondrial ribosomal protein L54
|
|
chr2_-_203103214
|
1.011
|
NM_001005781
NM_001005782
NM_003352
|
SUMO1
|
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae)
|
|
chr3_-_148939784
|
1.005
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr10_+_5005453
|
1.004
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr8_-_72274466
|
0.997
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr21_-_27423423
|
0.992
|
|
|
|
|
chr19_+_49259343
|
0.988
|
NM_019113
|
FGF21
|
fibroblast growth factor 21
|
|
chr4_-_186578122
|
0.979
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_69098502
|
0.978
|
NM_014482
|
BMP10
|
bone morphogenetic protein 10
|
|
chr1_+_50575465
|
0.977
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr21_-_27512668
|
0.972
|
NM_001136016
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr4_-_153273683
|
0.966
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr14_-_67826414
|
0.965
|
NM_015994
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D
|
|
chr20_-_55934877
|
0.965
|
NM_001190472
|
MTRNR2L3
|
MT-RNR2-like 3
|
|
chr4_+_111397181
|
0.948
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr5_-_32388595
|
0.938
|
|
ZFR
|
zinc finger RNA binding protein
|
|
chr5_+_86676620
|
0.918
|
|
|
|
|
chr20_+_2082527
|
0.895
|
NM_080836
|
STK35
|
serine/threonine kinase 35
|
|
chr5_+_125695787
|
0.892
|
NM_001146319
|
GRAMD3
|
GRAM domain containing 3
|
|
chr16_-_55867016
|
0.892
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr10_-_69455926
|
0.886
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr4_+_110354927
|
0.870
|
NM_001042734
NM_006323
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr1_+_84647342
|
0.843
|
NM_001242862
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr18_+_21032786
|
0.839
|
NM_003831
|
RIOK3
|
RIO kinase 3 (yeast)
|
|
chr14_+_62462540
|
0.839
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr10_-_94257470
|
0.811
|
NM_001165946
|
IDE
|
insulin-degrading enzyme
|
|
chr4_-_69434215
|
0.807
|
NM_001077
|
UGT2B15
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B15
UDP glucuronosyltransferase 2 family, polypeptide B17
|
|
chr18_+_21572736
|
0.803
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr8_+_104831415
|
0.797
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr13_-_25745778
|
0.796
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr9_-_112191105
|
0.792
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr1_+_248084319
|
0.790
|
NM_001005522
|
OR2T8
|
olfactory receptor, family 2, subfamily T, member 8
|
|
chrX_+_118108595
|
0.787
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr8_+_17396285
|
0.775
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr1_+_232941111
|
0.761
|
|
KIAA1383
|
KIAA1383
|
|
chr8_+_22210311
|
0.749
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr4_-_156787377
|
0.742
|
NM_017419
|
ACCN5
|
amiloride-sensitive cation channel 5, intestinal
|
|
chr4_+_110354971
|
0.737
|
|
SEC24B
|
SEC24 family, member B (S. cerevisiae)
|
|
chr5_+_161277702
|
0.728
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr10_+_5005685
|
0.726
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr17_+_57743498
|
0.723
|
|
CLTC
|
clathrin, heavy chain (Hc)
|
|
chr10_-_5045967
|
0.720
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr18_+_32621299
|
0.719
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr5_-_52405287
|
0.718
|
NM_004531
|
MOCS2
|
molybdenum cofactor synthesis 2
|
|
chr8_+_27179918
|
0.716
|
NM_004103
|
PTK2B
|
PTK2B protein tyrosine kinase 2 beta
|
|
chr2_-_133429069
|
0.714
|
NM_001077427
|
LYPD1
|
LY6/PLAUR domain containing 1
|
|
chr16_-_48266181
|
0.707
|
NM_033151
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11
|
|
chr5_-_137848517
|
0.707
|
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr17_-_685385
|
0.704
|
NM_016080
|
GLOD4
|
glyoxalase domain containing 4
|
|
chrX_+_49832214
|
0.703
|
NM_000084
|
CLCN5
|
chloride channel 5
|
|
chr8_+_94929991
|
0.701
|
NM_001161778
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1
|
|
chr19_-_6720585
|
0.685
|
NM_000064
|
C3
|
complement component 3
|
|
chr12_-_16757944
|
0.682
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr1_+_40839368
|
0.667
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr8_+_42396716
|
0.639
|
NM_138436
NM_001135674
NM_001135675
|
C8orf40
|
chromosome 8 open reading frame 40
|
|
chr1_-_109935869
|
0.638
|
NM_001205228
|
SORT1
|
sortilin 1
|
|
chr11_-_31014232
|
0.629
|
NM_020869
|
DCDC5
|
doublecortin domain containing 5
|
|
chr3_-_74570262
|
0.629
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr21_-_27253197
|
0.629
|
|
APP
|
amyloid beta (A4) precursor protein
|