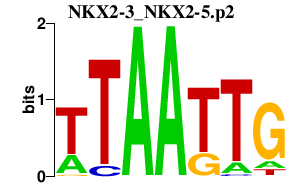
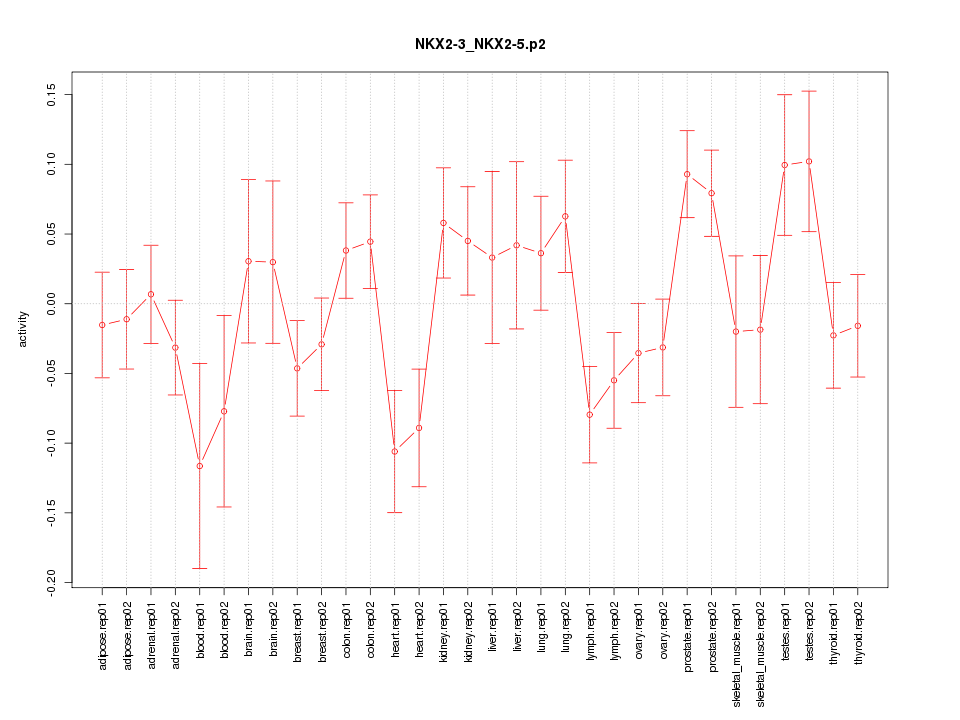
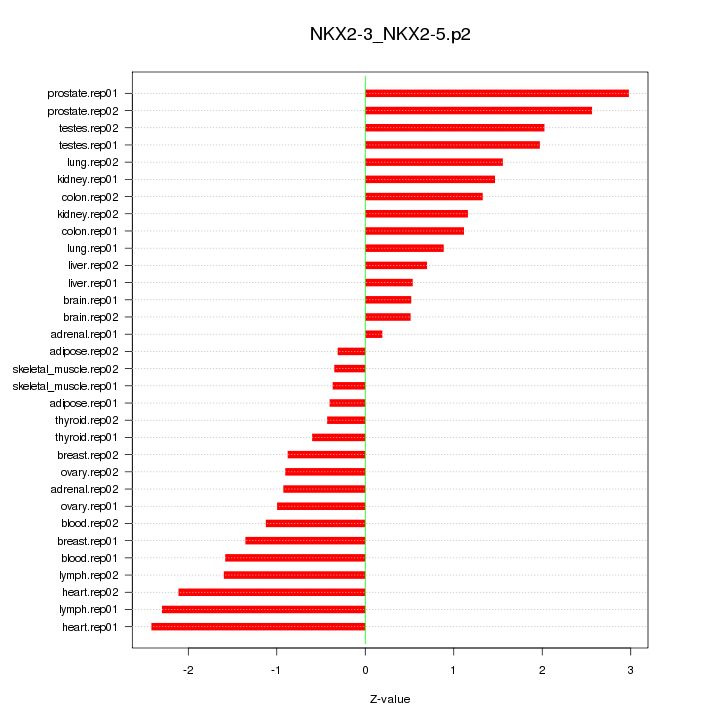
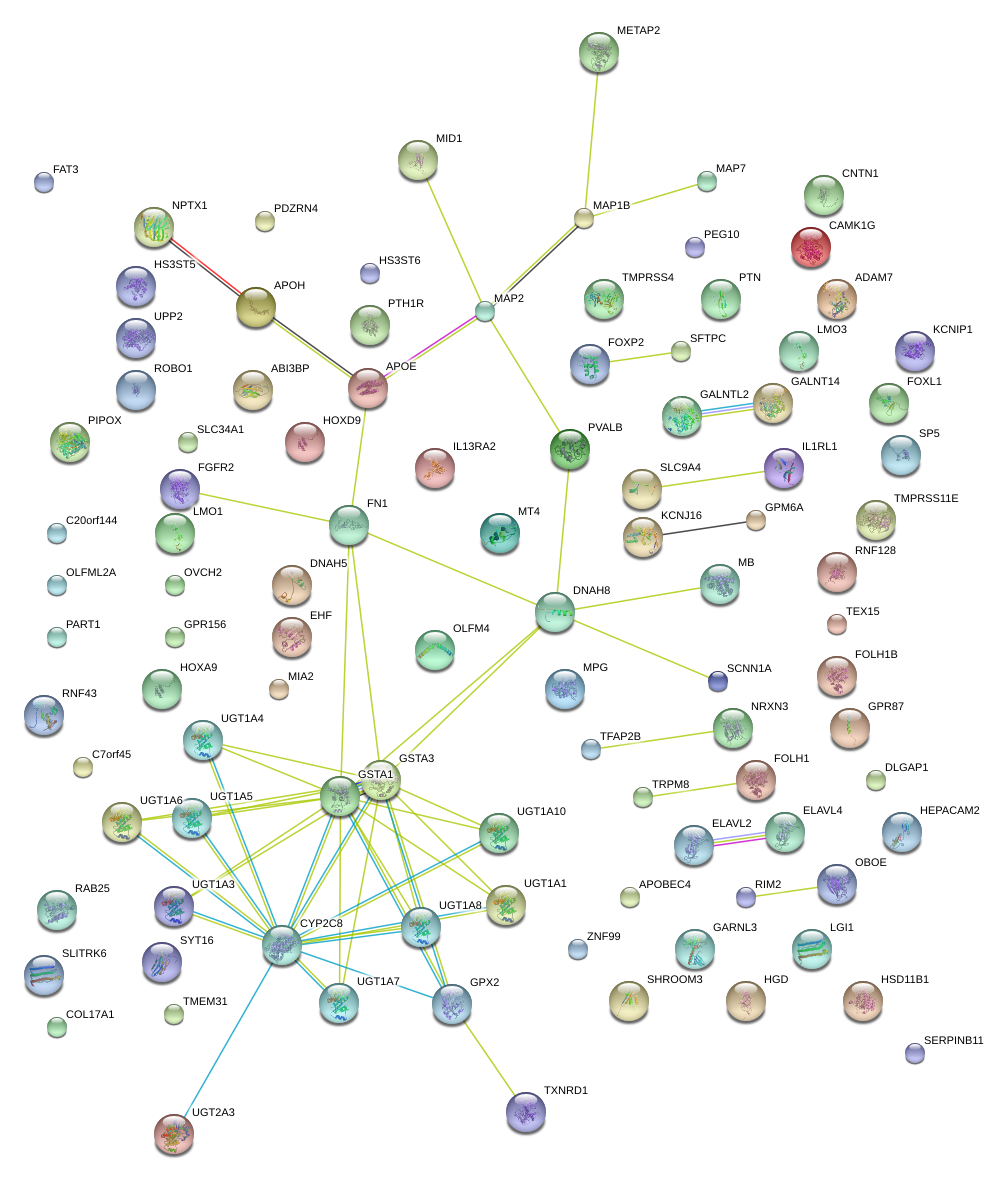
|
chr12_+_41831484
|
7.234
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr7_+_94297448
|
6.140
|
|
PEG10
|
paternally expressed 10
|
|
chr5_+_71502700
|
3.795
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr17_+_68071386
|
3.496
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr12_-_16760935
|
3.453
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_13944588
|
3.310
|
NM_001369
|
DNAH5
|
dynein, axonemal, heavy chain 5
|
|
chr13_-_86373328
|
3.221
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr8_+_104831415
|
3.197
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chrX_+_105937067
|
3.167
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr2_+_102953716
|
3.115
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr19_+_45409003
|
3.109
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr9_+_130026755
|
3.071
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr12_+_41086189
|
3.050
|
NM_001843
NM_175038
|
CNTN1
|
contactin 1
|
|
chr2_+_234826042
|
2.893
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr3_-_151034514
|
2.862
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr7_-_27205123
|
2.829
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_27205075
|
2.821
|
|
HOXA9
|
homeobox A9
|
|
chr12_-_6484389
|
2.802
|
NM_001159576
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr8_+_24298442
|
2.768
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr2_-_216245573
|
2.720
|
|
FN1
|
fibronectin 1
|
|
chr10_-_123356158
|
2.690
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr7_-_27205148
|
2.684
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr13_+_99058147
|
2.663
|
|
|
|
|
chr10_-_105845619
|
2.629
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr12_-_6484904
|
2.607
|
NM_001038
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr1_+_156030965
|
2.606
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr1_+_50574601
|
2.605
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_6484557
|
2.543
|
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr2_+_103089761
|
2.531
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr3_-_100712248
|
2.472
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr8_-_30706532
|
2.443
|
NM_031271
|
TEX15
|
testis expressed 15
|
|
chr17_-_78450247
|
2.409
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr6_-_136847078
|
2.382
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chr7_-_137028533
|
2.346
|
NM_002825
|
PTN
|
pleiotrophin
|
|
chr13_+_53602829
|
2.301
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr3_-_120401401
|
2.295
|
NM_000187
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr11_+_34642651
|
2.264
|
|
EHF
|
ets homologous factor
|
|
chr3_-_120400957
|
2.257
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr14_+_62462540
|
2.181
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chrX_+_102965836
|
2.141
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr17_+_27370155
|
2.081
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr14_+_39703119
|
2.079
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr7_+_114055048
|
2.075
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr3_-_79817058
|
2.067
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr10_-_105845556
|
2.059
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr2_+_234601511
|
2.058
|
NM_001072
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr4_-_69817478
|
2.043
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr4_-_176733901
|
2.039
|
|
GPM6A
|
glycoprotein M6A
|
|
chr3_-_119963141
|
2.038
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr10_-_96829074
|
1.986
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr5_+_169780880
|
1.975
|
NM_001034838
|
KCNIP1
|
Kv channel interacting protein 1
|
|
chr2_+_171571800
|
1.967
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr4_+_69313166
|
1.963
|
NM_014058
|
TMPRSS11E
|
transmembrane protease, serine 11E
|
|
chr5_+_59784004
|
1.941
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr1_+_209878135
|
1.919
|
NM_005525
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1
|
|
chr18_+_61370193
|
1.913
|
NM_080475
|
SERPINB11
|
serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene)
|
|
chr5_+_176811431
|
1.892
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chrX_-_10544852
|
1.860
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_31352489
|
1.859
|
NM_001253827
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14)
|
|
chr2_+_210444364
|
1.838
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr3_+_46919235
|
1.837
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr7_+_129847699
|
1.782
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr10_+_95517565
|
1.777
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr14_+_78870092
|
1.774
|
NM_004796
|
NRXN3
|
neurexin 3
|
|
chr16_+_86612114
|
1.750
|
NM_005250
|
FOXL1
|
forkhead box L1
|
|
chr12_-_16759430
|
1.749
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_56494889
|
1.748
|
|
RNF43
|
ring finger protein 43
|
|
chr3_+_16216155
|
1.724
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chr14_-_65409445
|
1.711
|
NM_002083
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal)
|
|
chr20_+_32250089
|
1.709
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr11_+_92085261
|
1.704
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chrX_-_114252132
|
1.687
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr11_+_34642587
|
1.673
|
NM_001206615
NM_012153
|
EHF
|
ets homologous factor
|
|
chr17_-_64225496
|
1.673
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr7_-_92855780
|
1.621
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr17_+_68100994
|
1.619
|
NM_170742
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr6_-_114384008
|
1.592
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr18_-_3845295
|
1.585
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr22_-_37215516
|
1.582
|
NM_002854
|
PVALB
|
parvalbumin
|
|
chr11_+_89392464
|
1.578
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr2_+_158851690
|
1.559
|
NM_001135098
|
UPP2
|
uridine phosphorylase 2
|
|
chr12_-_16757944
|
1.524
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_+_117947785
|
1.520
|
|
TMPRSS4
|
transmembrane protease, serine 4
|
|
chr6_-_52774460
|
1.519
|
NM_000847
|
GSTA3
|
glutathione S-transferase alpha 3
|
|
chr3_+_159943252
|
1.507
|
NM_001168214
|
C3orf80
|
chromosome 3 open reading frame 80
|
|
chr1_-_183622447
|
1.493
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr11_-_7727940
|
1.489
|
NM_198185
|
OVCH2
|
ovochymase 2 (gene/pseudogene)
|
|
chr1_+_209757044
|
1.482
|
NM_020439
|
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG
|
|
chr2_+_234627423
|
1.480
|
NM_007120
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4
|
|
chr6_+_50786438
|
1.468
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr4_+_77356252
|
1.464
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chrX_+_111119278
|
1.464
|
NM_001195576
NM_001195578
|
LOC100329135
|
uncharacterized LOC100329135
|
|
chr8_-_86290333
|
1.457
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr1_+_50574589
|
1.444
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr22_-_38506532
|
1.442
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr12_-_24103842
|
1.441
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr7_-_14029641
|
1.433
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr4_-_110723127
|
1.431
|
|
CFI
|
complement factor I
|
|
chr4_-_111563278
|
1.429
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr13_+_46276445
|
1.429
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr9_-_27297106
|
1.426
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr14_-_38064440
|
1.421
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr13_-_61989654
|
1.416
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr4_-_87281374
|
1.409
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr16_-_11375094
|
1.400
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr4_-_138453628
|
1.394
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr2_-_79386878
|
1.394
|
NM_138938
NM_002580
NM_138937
|
REG3A
|
regenerating islet-derived 3 alpha
|
|
chr2_-_183387063
|
1.371
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr5_+_34687663
|
1.361
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr3_-_170303774
|
1.361
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (orphan transporter), member 14
|
|
chr12_-_49488570
|
1.359
|
NM_021044
|
DHH
|
desert hedgehog
|
|
chr12_-_16759420
|
1.355
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr19_-_48389571
|
1.344
|
NM_003167
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr4_-_176734181
|
1.339
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr20_-_29978285
|
1.333
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr5_+_40909558
|
1.326
|
NM_000587
|
C7
|
complement component 7
|
|
chr12_-_24103953
|
1.311
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr21_-_33957746
|
1.310
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr1_-_57045235
|
1.298
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_87804773
|
1.292
|
NM_000735
NM_001252383
|
CGA
|
glycoprotein hormones, alpha polypeptide
|
|
chr5_+_40909617
|
1.290
|
|
C7
|
complement component 7
|
|
chr1_+_50575465
|
1.284
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_16759939
|
1.279
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr16_-_15835382
|
1.258
|
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr13_+_78109808
|
1.253
|
NM_001160706
NM_003843
NM_144777
|
SCEL
|
sciellin
|
|
chr5_+_68710938
|
1.251
|
NM_001038603
NM_001244734
|
MARVELD2
|
MARVEL domain containing 2
|
|
chr5_+_152870083
|
1.248
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chr1_+_35246789
|
1.238
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr19_-_48389514
|
1.232
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr3_-_187009469
|
1.231
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr9_-_117265452
|
1.226
|
NM_001083885
|
DFNB31
|
deafness, autosomal recessive 31
|
|
chr4_+_69681709
|
1.221
|
NM_001075
NM_001144767
|
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10
|
|
chr6_-_49931817
|
1.220
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr2_+_211458078
|
1.217
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr12_-_88423145
|
1.210
|
NM_152589
|
C12orf50
|
chromosome 12 open reading frame 50
|
|
chr1_+_50569581
|
1.203
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chrX_+_22050920
|
1.182
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chrX_-_15288172
|
1.181
|
NM_001031739
NM_001168530
NM_024087
|
ASB9
|
ankyrin repeat and SOCS box containing 9
|
|
chr20_+_44350987
|
1.180
|
NM_178455
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4
|
|
chr7_-_121944485
|
1.179
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr9_-_23826062
|
1.177
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr6_+_134758840
|
1.171
|
|
LOC154092
|
uncharacterized LOC154092
|
|
chr8_-_82373757
|
1.170
|
NM_001080526
|
FABP9
|
fatty acid binding protein 9, testis
|
|
chr12_+_49741040
|
1.168
|
NM_024902
|
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr1_-_57045129
|
1.160
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_52645430
|
1.158
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr13_+_96085852
|
1.154
|
NM_001160100
NM_182848
|
CLDN10
|
claudin 10
|
|
chr7_-_107880492
|
1.152
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr1_+_63063186
|
1.149
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr13_-_36429798
|
1.143
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr4_-_87374282
|
1.140
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr11_+_101983170
|
1.139
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr3_-_149095413
|
1.132
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr2_+_158114109
|
1.127
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr3_+_85775631
|
1.125
|
NM_153184
|
CADM2
|
cell adhesion molecule 2
|
|
chr9_+_118916069
|
1.120
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr17_-_42908178
|
1.115
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr17_+_56833231
|
1.111
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr12_-_91505215
|
1.110
|
|
LUM
|
lumican
|
|
chrX_-_10851808
|
1.088
|
NM_033290
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr5_-_35938736
|
1.083
|
NM_001042625
NM_144647
|
CAPSL
|
calcyphosine-like
|
|
chr9_-_95166827
|
1.081
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr4_-_152149042
|
1.079
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr1_-_57045214
|
1.059
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr2_+_189839078
|
1.052
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chrX_+_30248552
|
1.049
|
NM_002365
|
MAGEB3
|
melanoma antigen family B, 3
|
|
chr12_+_52056547
|
1.043
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr12_-_16759733
|
1.033
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr3_-_74570262
|
1.032
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr1_+_147374945
|
1.028
|
NM_005267
|
GJA8
|
gap junction protein, alpha 8, 50kDa
|
|
chr4_+_75174189
|
1.025
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr1_-_116383692
|
1.021
|
NM_005599
|
NHLH2
|
nescient helix loop helix 2
|
|
chr12_-_16759531
|
1.020
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_-_85437457
|
1.020
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr7_-_41742666
|
1.016
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr19_-_36304200
|
1.015
|
NM_021232
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr2_+_48796158
|
1.013
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chrX_+_50027539
|
0.995
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr5_-_137475076
|
0.990
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr10_+_101542460
|
0.985
|
NM_000392
|
ABCC2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2
|
|
chr16_-_20416032
|
0.984
|
NM_174924
|
PDILT
|
protein disulfide isomerase-like, testis expressed
|
|
chr4_+_30721865
|
0.982
|
NM_032456
NM_001173523
NM_002589
NM_032457
|
PCDH7
|
protocadherin 7
|
|
chr1_+_68150858
|
0.973
|
NM_001199741
NM_001199742
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr15_-_69707358
|
0.972
|
|
LOC145694
|
uncharacterized LOC145694
|
|
chr5_-_148758787
|
0.969
|
NM_014443
|
IL17B
|
interleukin 17B
|
|
chr3_+_2933898
|
0.964
|
NM_001206956
NM_175613
|
CNTN4
|
contactin 4
|
|
chr5_+_31193852
|
0.964
|
|
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney)
|
|
chr16_-_20338808
|
0.960
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr21_+_19617145
|
0.959
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr9_-_104357202
|
0.958
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr10_+_124670216
|
0.956
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr4_+_155484131
|
0.947
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chrX_-_55057408
|
0.946
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr4_+_78432905
|
0.944
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr2_+_171571936
|
0.944
|
|
|
|
|
chr18_-_40857245
|
0.940
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr17_-_38956146
|
0.937
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr1_-_54199876
|
0.936
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr20_+_62795826
|
0.935
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr12_-_52914080
|
0.935
|
NM_000424
|
KRT5
|
keratin 5
|