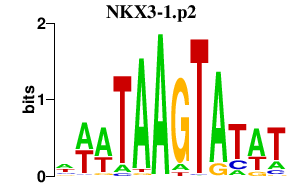
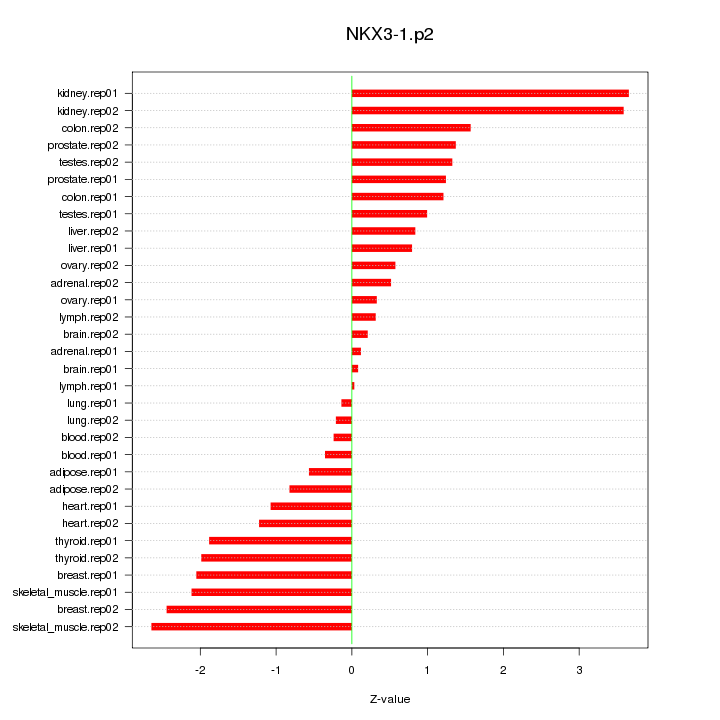
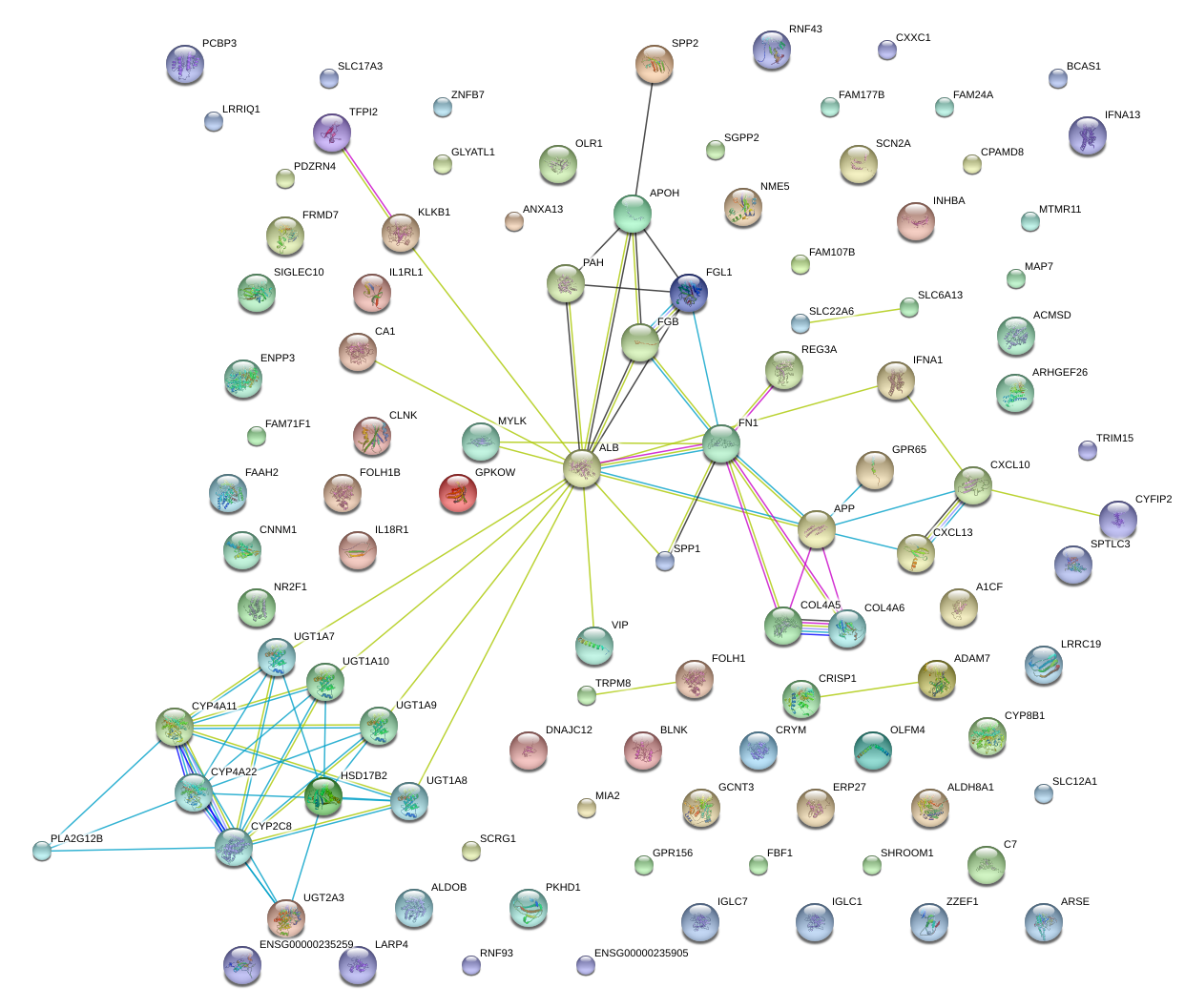
|
chr9_-_104198045
|
7.424
|
NM_000035
|
ALDOB
|
aldolase B, fructose-bisphosphate
|
|
chr4_+_88896838
|
5.967
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_88896801
|
5.608
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_88896868
|
5.508
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr4_+_88896899
|
5.039
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr6_+_131958441
|
4.936
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr12_+_41831484
|
4.509
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr17_-_64225496
|
4.304
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chrX_-_131261871
|
3.951
|
NM_194277
|
FRMD7
|
FERM domain containing 7
|
|
chr10_+_101088794
|
3.844
|
NM_020348
|
CNNM1
|
cyclin M1
|
|
chr4_+_74269971
|
3.426
|
NM_000477
|
ALB
|
albumin
|
|
chr17_-_56492938
|
3.382
|
|
RNF43
|
ring finger protein 43
|
|
chr5_+_156696351
|
3.293
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr3_-_42917191
|
3.069
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr2_+_135596185
|
2.800
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr8_-_124749573
|
2.739
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr5_+_92918924
|
2.700
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr15_+_59904090
|
2.699
|
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr2_+_102953716
|
2.674
|
NM_003856
|
IL1RL1
|
interleukin 1 receptor-like 1
|
|
chr12_-_103311380
|
2.632
|
NM_000277
|
PAH
|
phenylalanine hydroxylase
|
|
chr10_-_74714479
|
2.622
|
NM_032562
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr12_-_15091441
|
2.489
|
NM_152321
|
ERP27
|
endoplasmic reticulum protein 27
|
|
chr6_-_136847078
|
2.464
|
NM_001198608
NM_001198609
NM_001198611
|
MAP7
|
microtubule-associated protein 7
|
|
chrX_-_2882288
|
2.441
|
NM_000047
|
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1)
|
|
chr10_-_69597768
|
2.412
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr12_+_85430098
|
2.394
|
NM_001079910
|
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1
|
|
chr1_-_47407094
|
2.374
|
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11
|
|
chr9_-_27005649
|
2.333
|
NM_022901
|
LRRC19
|
leucine rich repeat containing 19
|
|
chr5_-_137475076
|
2.324
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr14_+_39703119
|
2.288
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr11_+_58695101
|
2.261
|
NM_001220494
|
GLYATL1
|
glycine-N-acyltransferase-like 1
|
|
chr2_+_102927961
|
2.243
|
NM_016232
|
IL18R1
IL1RL1
|
interleukin 18 receptor 1
interleukin 1 receptor-like 1
|
|
chr11_-_62752370
|
2.214
|
NM_004790
NM_153276
NM_153277
NM_153278
|
SLC22A6
|
solute carrier family 22 (organic anion transporter), member 6
|
|
chr15_+_59903980
|
2.200
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr13_+_53602829
|
2.142
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr6_-_25874439
|
2.085
|
NM_001098486
NM_006632
|
SLC17A3
|
solute carrier family 17 (sodium phosphate), member 3
|
|
chr4_-_174320616
|
2.049
|
NM_007281
|
SCRG1
|
stimulator of chondrogenesis 1
|
|
chr20_-_52687303
|
2.019
|
NM_003657
|
BCAS1
|
breast carcinoma amplified sequence 1
|
|
chr8_-_17743062
|
2.010
|
|
FGL1
|
fibrinogen-like 1
|
|
chrX_+_107683015
|
1.979
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chr3_-_119963141
|
1.961
|
NM_001168271
NM_153002
|
GPR156
|
G protein-coupled receptor 156
|
|
chr2_+_234590583
|
1.942
|
NM_019077
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7
|
|
chr1_-_47407142
|
1.935
|
NM_000778
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11
|
|
chr1_-_149908098
|
1.915
|
NM_181873
|
MTMR11
|
myotubularin related protein 11
|
|
chr6_-_51952368
|
1.892
|
NM_138694
NM_170724
|
PKHD1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)
|
|
chr19_-_51920841
|
1.881
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr11_+_89392464
|
1.869
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr1_+_222910557
|
1.844
|
NM_207468
|
FAM177B
|
family with sequence similarity 177, member B
|
|
chr12_-_371994
|
1.841
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr2_+_234959345
|
1.834
|
NM_006944
|
SPP2
|
secreted phosphoprotein 2, 24kDa
|
|
chr10_-_52645430
|
1.834
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr6_-_49834188
|
1.815
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr7_-_41742666
|
1.743
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr6_-_135271206
|
1.717
|
NM_001193480
NM_022568
NM_170771
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1
|
|
chr15_+_48498497
|
1.713
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr14_+_88471467
|
1.707
|
NM_003608
|
GPR65
|
G protein-coupled receptor 65
|
|
chr21_+_47269874
|
1.698
|
NM_001130141
NM_020528
|
PCBP3
|
poly(rC) binding protein 3
|
|
chr16_-_21314371
|
1.679
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr6_+_30130982
|
1.660
|
NM_033229
|
TRIM15
|
tripartite motif containing 15
|
|
chr6_+_153071931
|
1.630
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr21_-_27253197
|
1.626
|
|
APP
|
amyloid beta (A4) precursor protein
|
|
chr7_+_45933096
|
1.624
|
|
|
|
|
chr16_+_82068830
|
1.595
|
NM_002153
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr10_-_98031154
|
1.575
|
NM_001114094
NM_013314
|
BLNK
|
B-cell linker
|
|
chr2_+_166095911
|
1.557
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr9_+_21440452
|
1.543
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr4_-_69817478
|
1.539
|
NM_024743
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3
|
|
chr4_+_155484131
|
1.525
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr7_+_128349111
|
1.522
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr16_+_82068958
|
1.520
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr10_+_124670216
|
1.519
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr10_-_14816895
|
1.472
|
NM_031453
|
FAM107B
|
family with sequence similarity 107, member B
|
|
chr12_-_10324675
|
1.467
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr4_+_78432905
|
1.443
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr7_-_93519990
|
1.440
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr8_-_86253863
|
1.405
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr2_-_79386878
|
1.400
|
NM_138938
NM_002580
NM_138937
|
REG3A
|
regenerating islet-derived 3 alpha
|
|
chr16_+_82068910
|
1.375
|
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2
|
|
chr12_+_50869531
|
1.374
|
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr10_-_96829074
|
1.351
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chrX_-_107682600
|
1.345
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr4_+_187148624
|
1.344
|
NM_000892
|
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1
|
|
chr5_+_121495870
|
1.341
|
NM_001195535
|
LOC100505841
|
zinc finger protein 474-like
|
|
chr16_+_4845265
|
1.338
|
NM_001253793
NM_001253794
|
LOC440335
|
uncharacterized LOC440335
|
|
chr20_+_12989616
|
1.332
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr4_-_10686314
|
1.301
|
NM_052964
|
CLNK
|
cytokine-dependent hematopoietic cell linker
|
|
chrX_+_57313109
|
1.296
|
NM_174912
|
FAAH2
|
fatty acid amide hydrolase 2
|
|
chr3_+_153838791
|
1.295
|
NM_001251962
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr2_-_216248184
|
1.295
|
|
FN1
|
fibronectin 1
|
|
chr3_-_123339178
|
1.286
|
|
MYLK
|
myosin light chain kinase
|
|
chr8_+_24298442
|
1.286
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr2_+_234826042
|
1.270
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr5_-_132161863
|
1.258
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr5_+_40909558
|
1.234
|
NM_000587
|
C7
|
complement component 7
|
|
chr6_-_49712055
|
1.219
|
NM_001190986
NM_006061
|
CRISP3
|
cysteine-rich secretory protein 3
|
|
chr5_+_40909617
|
1.200
|
|
C7
|
complement component 7
|
|
chr7_-_123673486
|
1.198
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr3_-_93747453
|
1.193
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr14_+_94492683
|
1.184
|
NM_023112
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr3_-_120400957
|
1.175
|
|
HGD
|
homogentisate 1,2-dioxygenase
|
|
chr6_-_76782204
|
1.171
|
NM_001563
|
IMPG1
|
interphotoreceptor matrix proteoglycan 1
|
|
chr5_+_126988706
|
1.150
|
NM_001127385
|
CTXN3
|
cortexin 3
|
|
chr3_+_142375738
|
1.130
|
NM_001172312
|
PLS1
|
plastin 1
|
|
chr12_+_57157091
|
1.127
|
NM_003725
|
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse)
|
|
chr19_-_53794874
|
1.122
|
NM_033341
|
BIRC8
|
baculoviral IAP repeat containing 8
|
|
chr12_-_80084742
|
1.105
|
NM_002583
|
PAWR
|
PRKC, apoptosis, WT1, regulator
|
|
chr2_+_166150340
|
1.104
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr18_+_72167021
|
1.104
|
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family)
|
|
chr9_+_90112754
|
1.103
|
NM_004938
|
DAPK1
|
death-associated protein kinase 1
|
|
chr9_+_127119114
|
1.102
|
|
|
|
|
chr3_-_194072004
|
1.094
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr8_+_24151579
|
1.083
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr12_-_81763127
|
1.081
|
NM_001220479
NM_001220480
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr7_-_92855780
|
1.074
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr5_+_40909487
|
1.073
|
|
C7
|
complement component 7
|
|
chr14_+_95078713
|
1.073
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr10_-_118680978
|
1.072
|
|
KIAA1598
|
KIAA1598
|
|
chr6_-_49931817
|
1.071
|
NM_001037499
|
DEFB114
|
defensin, beta 114
|
|
chr17_-_38859976
|
1.058
|
NM_019016
|
KRT24
|
keratin 24
|
|
chr2_-_183387063
|
1.050
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr9_-_136604894
|
1.050
|
NM_007101
|
SARDH
|
sarcosine dehydrogenase
|
|
chr8_-_15095791
|
1.049
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chrX_-_100548044
|
1.044
|
NM_024885
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
|
|
chr3_+_108855560
|
1.040
|
|
FLJ22763
|
uncharacterized LOC401081
|
|
chr1_+_171217632
|
1.029
|
NM_002021
|
FMO1
|
flavin containing monooxygenase 1
|
|
chr5_+_140261792
|
1.022
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr8_-_17752847
|
1.022
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr3_-_146213721
|
1.018
|
NM_001199979
|
PLSCR2
|
phospholipid scramblase 2
|
|
chr10_+_90346509
|
1.005
|
NM_001010939
|
LIPJ
|
lipase, family member J
|
|
chr18_-_64271215
|
1.003
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr19_-_1021122
|
0.985
|
NM_001033026
NM_033420
|
C19orf6
|
chromosome 19 open reading frame 6
|
|
chr21_-_34185943
|
0.972
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_+_57320442
|
0.967
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chrX_-_114252132
|
0.967
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr1_+_207262583
|
0.957
|
NM_000716
NM_001017364
NM_001017367
|
C4BPB
|
complement component 4 binding protein, beta
|
|
chr4_+_186350543
|
0.950
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr3_-_172858959
|
0.930
|
NM_031955
|
SPATA16
|
spermatogenesis associated 16
|
|
chr11_-_128712340
|
0.927
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr9_-_125941304
|
0.926
|
|
STRBP
|
spermatid perinuclear RNA binding protein
|
|
chr4_-_155312448
|
0.921
|
NM_017639
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr3_-_74570262
|
0.918
|
NM_020872
|
CNTN3
|
contactin 3 (plasmacytoma associated)
|
|
chr7_+_115863004
|
0.913
|
NM_152829
|
TES
|
testis derived transcript (3 LIM domains)
|
|
chr4_+_156824846
|
0.908
|
NM_005651
|
TDO2
|
tryptophan 2,3-dioxygenase
|
|
chr1_+_159557615
|
0.908
|
NM_001639
|
APCS
|
amyloid P component, serum
|
|
chr20_+_44330654
|
0.898
|
NM_172005
|
WFDC13
|
WAP four-disulfide core domain 13
|
|
chr3_+_46919235
|
0.897
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr17_-_7493389
|
0.897
|
NM_006942
|
SOX15
|
SRY (sex determining region Y)-box 15
|
|
chr14_+_94492747
|
0.896
|
|
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2
|
|
chr6_+_31895201
|
0.896
|
NM_000063
NM_001145903
|
C2
|
complement component 2
|
|
chr1_+_84630575
|
0.889
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr2_-_165551178
|
0.876
|
|
COBLL1
|
COBL-like 1
|
|
chr5_-_36301999
|
0.875
|
NM_001161429
NM_145000
|
RANBP3L
|
RAN binding protein 3-like
|
|
chr6_+_136172802
|
0.851
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr3_-_42917632
|
0.848
|
NM_004391
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr17_+_7788094
|
0.846
|
NM_001005271
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr17_+_61086897
|
0.839
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr1_-_112106562
|
0.836
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr1_+_101185195
|
0.836
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chrX_+_83116153
|
0.832
|
NM_021118
|
CYLC1
|
cylicin, basic protein of sperm head cytoskeleton 1
|
|
chr1_-_86043932
|
0.832
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_-_151770611
|
0.831
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr2_-_224866616
|
0.830
|
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr4_-_71532206
|
0.822
|
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides
|
|
chr4_+_75174189
|
0.821
|
NM_001013442
|
EPGN
|
epithelial mitogen homolog (mouse)
|
|
chr5_+_147691988
|
0.816
|
NM_032566
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative)
|
|
chr10_+_96698409
|
0.810
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr13_+_50025687
|
0.809
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr11_+_6942204
|
0.803
|
NM_001004684
|
OR2D3
|
olfactory receptor, family 2, subfamily D, member 3
|
|
chr7_-_107880492
|
0.801
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr20_+_43835637
|
0.800
|
NM_003007
|
SEMG1
|
semenogelin I
|
|
chrX_+_114827818
|
0.786
|
NM_001136025
NM_001172335
|
PLS3
|
plastin 3
|
|
chr4_+_166131170
|
0.782
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chrX_-_13835012
|
0.775
|
|
GPM6B
|
glycoprotein M6B
|
|
chr4_+_74270832
|
0.775
|
|
ALB
|
albumin
|
|
chr2_-_27718059
|
0.771
|
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr20_+_44350987
|
0.768
|
NM_178455
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4
|
|
chr10_-_123353330
|
0.768
|
NM_001144913
NM_001144914
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr20_-_29978285
|
0.764
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr2_-_88285308
|
0.760
|
NM_001024457
|
RGPD1
|
RANBP2-like and GRIP domain containing 1
|
|
chr4_+_159442865
|
0.755
|
NM_001253727
NM_001253728
NM_001253729
NM_001253730
NM_001253732
NM_001253733
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr4_+_88529680
|
0.751
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr2_+_173686314
|
0.751
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr6_+_31895401
|
0.743
|
|
C2
|
complement component 2
|
|
chr5_+_140235594
|
0.742
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr6_-_49989647
|
0.736
|
NM_001037497
NM_001037728
|
DEFB110
|
defensin, beta 110 locus
|
|
chr12_-_13248581
|
0.727
|
NM_001206843
NM_001206845
NM_031289
NM_153823
|
GSG1
|
germ cell associated 1
|
|
chr9_+_90112419
|
0.726
|
|
DAPK1
|
death-associated protein kinase 1
|
|
chr2_+_103089761
|
0.725
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr1_-_202129705
|
0.722
|
|
PTPN7
|
protein tyrosine phosphatase, non-receptor type 7
|
|
chrX_+_105937067
|
0.719
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr15_+_94899163
|
0.718
|
NM_001159644
|
MCTP2
|
multiple C2 domains, transmembrane 2
|
|
chr2_-_233641225
|
0.716
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr2_-_27718098
|
0.713
|
NM_022823
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr2_-_27362277
|
0.709
|
NM_178553
|
C2orf53
|
chromosome 2 open reading frame 53
|
|
chr13_-_86373328
|
0.703
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr3_+_151591428
|
0.693
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr1_-_15911461
|
0.681
|
NM_024758
|
AGMAT
|
agmatine ureohydrolase (agmatinase)
|
|
chr6_+_27292539
|
0.678
|
NM_032030
|
FKSG83
|
FKSG83
|
|
chr2_+_207804277
|
0.677
|
NM_173077
|
CPO
|
carboxypeptidase O
|
|
chr1_+_196857143
|
0.677
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|