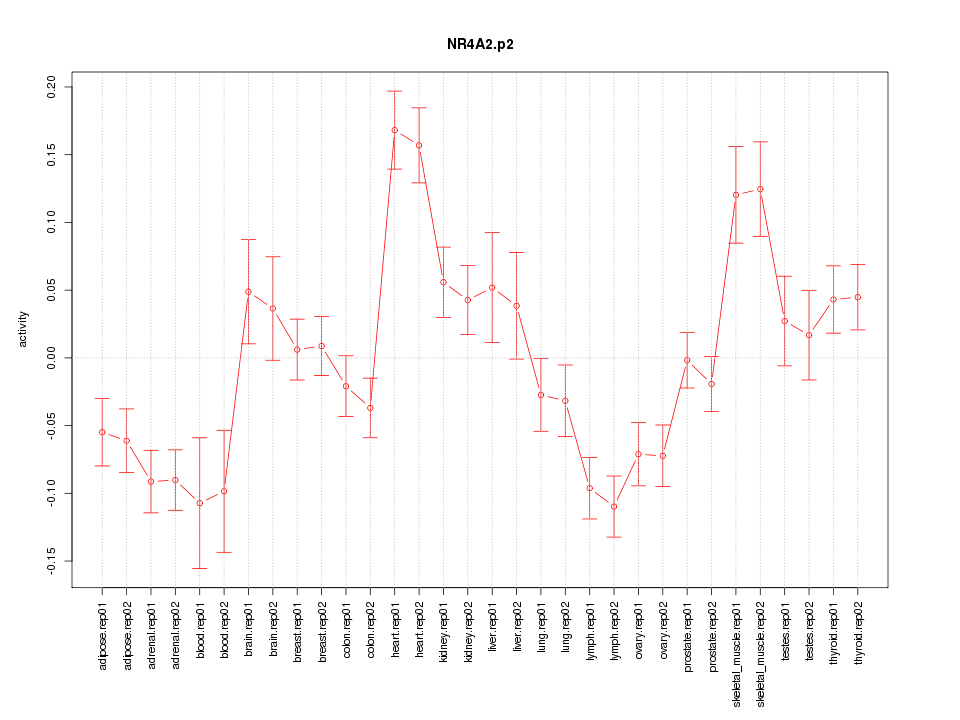
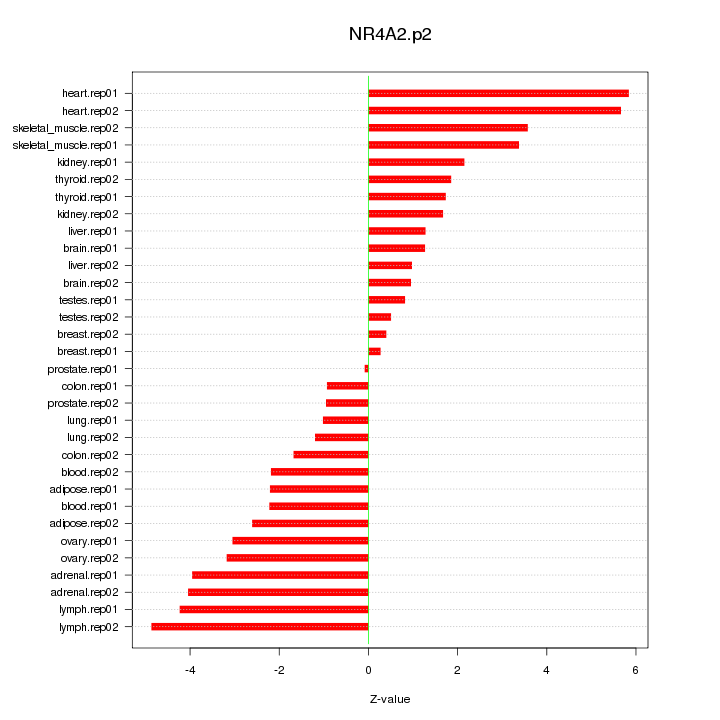
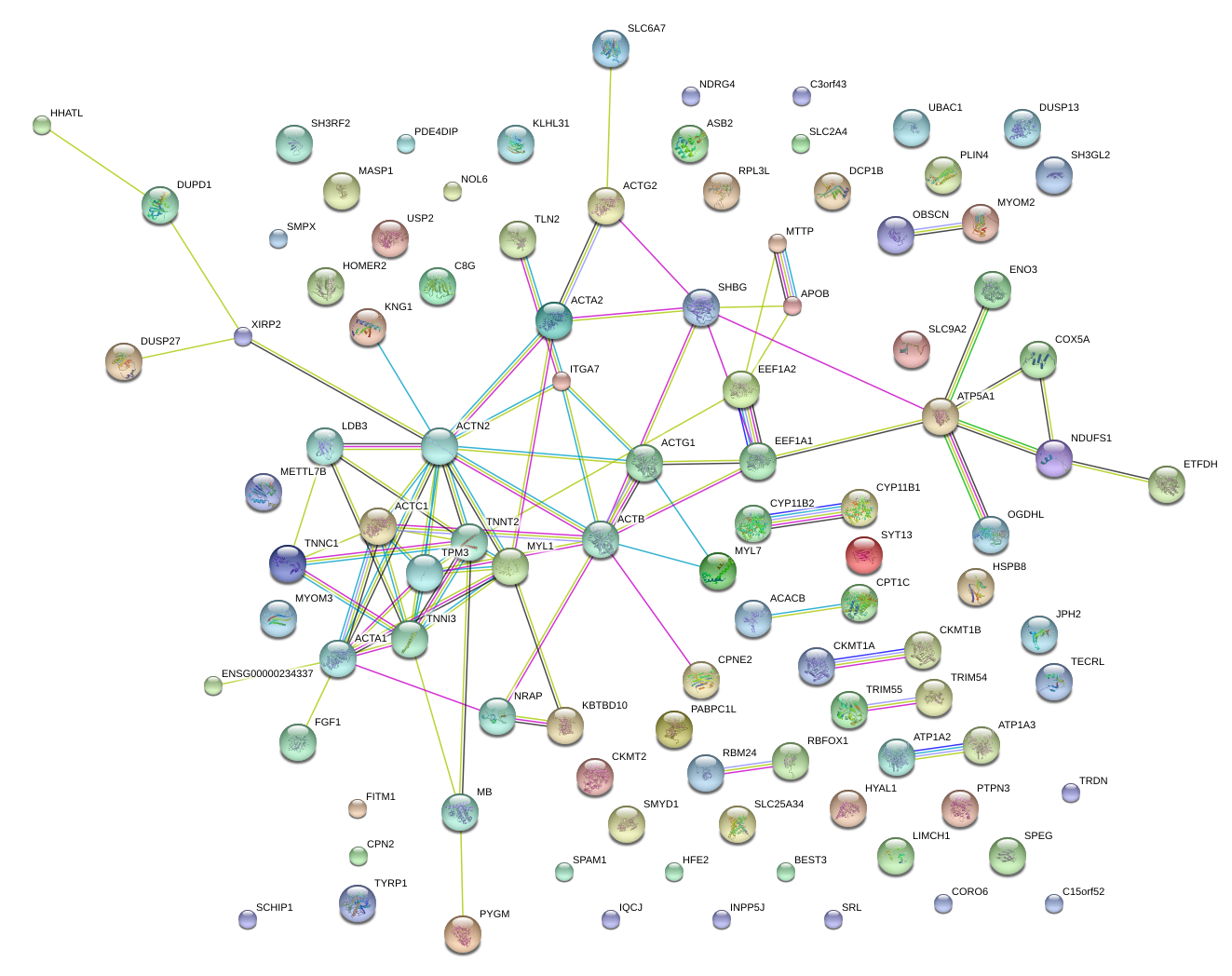
|
chr1_+_236849753
|
20.476
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr20_-_62130390
|
13.017
|
NM_001958
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2
|
|
chr5_+_80529138
|
10.506
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chrX_-_21776158
|
9.245
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr3_-_42744318
|
9.195
|
NM_020707
|
HHATL
|
hedgehog acyltransferase-like
|
|
chr16_-_4292052
|
8.263
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr2_+_88367298
|
8.255
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr22_-_36019363
|
8.137
|
NM_203377
|
MB
|
myoglobin
|
|
chr8_+_67039130
|
7.889
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr17_+_4854383
|
7.423
|
NM_001193503
NM_001976
NM_053013
|
ENO3
|
enolase 3 (beta, muscle)
|
|
chr3_-_194072004
|
7.194
|
NM_001080513
|
CPN2
|
carboxypeptidase N, polypeptide 2
|
|
chr11_-_64528186
|
6.993
|
NM_001164716
NM_005609
|
PYGM
|
phosphorylase, glycogen, muscle
|
|
chr6_-_53530505
|
6.727
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr1_-_229569839
|
6.238
|
NM_001100
|
ACTA1
|
actin, alpha 1, skeletal muscle
|
|
chr1_-_154164551
|
6.086
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr4_+_41614725
|
6.000
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_65275000
|
5.876
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr3_-_47957475
|
5.831
|
|
|
|
|
chr5_+_145316120
|
5.793
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr2_+_170366211
|
5.787
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr16_+_6069094
|
5.699
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr4_+_41614933
|
5.654
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr1_+_228395740
|
5.533
|
NM_001098623
NM_052843
|
OBSCN
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF
|
|
chr7_-_44180911
|
5.440
|
NM_021223
|
MYL7
|
myosin, light chain 7, regulatory
|
|
chr3_+_159557649
|
5.246
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_56075418
|
5.204
|
|
METTL7B
|
methyltransferase like 7B
|
|
chr6_-_123957941
|
5.151
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr4_+_41614911
|
5.074
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_-_196242190
|
4.989
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr12_+_56075329
|
4.954
|
NM_152637
|
METTL7B
|
methyltransferase like 7B
|
|
chr19_-_4517715
|
4.789
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr2_+_168043792
|
4.775
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr3_+_186435097
|
4.771
|
NM_000893
NM_001102416
NM_001166451
|
KNG1
|
kininogen 1
|
|
chr6_+_17282931
|
4.681
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_+_220299699
|
4.642
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr11_-_119234628
|
4.575
|
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr1_-_201346804
|
4.511
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr17_+_7533452
|
4.493
|
NM_001040
NM_001146279
NM_001146280
NM_001146281
|
SHBG
|
sex hormone-binding globulin
|
|
chr1_-_144994908
|
4.476
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr11_-_45307617
|
4.472
|
NM_001247987
NM_020826
|
SYT13
|
synaptotagmin XIII
|
|
chr2_-_207024103
|
4.444
|
NM_001199981
NM_001199982
NM_001199983
NM_005006
|
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase)
|
|
chr20_-_42816217
|
4.150
|
NM_020433
NM_175913
|
JPH2
|
junctophilin 2
|
|
chr22_+_31518923
|
4.143
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr12_+_119616594
|
4.082
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr10_-_76859247
|
4.066
|
NM_016364
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr3_-_50340979
|
4.046
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr4_+_100495972
|
3.988
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr17_-_27948345
|
3.921
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr8_+_1993157
|
3.911
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr1_+_160085519
|
3.901
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr5_-_142077508
|
3.834
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_+_167064086
|
3.763
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr3_+_159557745
|
3.762
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_56106061
|
3.747
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr19_-_55668956
|
3.707
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr3_-_187009469
|
3.555
|
NM_001031849
NM_001879
NM_139125
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor)
|
|
chr10_+_88428167
|
3.530
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr2_+_103236147
|
3.530
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr14_+_24600674
|
3.472
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr2_-_211168315
|
3.408
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr16_-_2004670
|
3.371
|
NM_005061
|
RPL3L
|
ribosomal protein L3-like
|
|
chr5_+_149569519
|
3.342
|
NM_014228
|
SLC6A7
|
solute carrier family 6 (neurotransmitter transporter, L-proline), member 7
|
|
chr15_-_40630272
|
3.305
|
|
C15orf52
|
chromosome 15 open reading frame 52
|
|
chr1_-_144994839
|
3.297
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr2_+_27505296
|
3.295
|
NM_032546
NM_187841
|
TRIM54
|
tripartite motif containing 54
|
|
chr9_+_139839697
|
3.293
|
NM_000606
|
C8G
|
complement component 8, gamma polypeptide
|
|
chr1_+_16062808
|
3.192
|
NM_207348
|
SLC25A34
|
solute carrier family 25, member 34
|
|
chr2_-_21225640
|
3.186
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr12_-_70093064
|
3.170
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr1_-_24434927
|
3.149
|
|
MYOM3
|
myomesin family, member 3
|
|
chr5_+_145317297
|
3.062
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr4_+_159593233
|
3.012
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr15_-_83621350
|
3.004
|
NM_004839
NM_199330
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr14_-_94442894
|
2.989
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr9_+_12693385
|
2.958
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr10_-_50970321
|
2.936
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr9_-_112191105
|
2.923
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr10_-_115423640
|
2.899
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr16_+_58534045
|
2.874
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr18_-_43678230
|
2.857
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr17_+_7184978
|
2.849
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr1_+_145413190
|
2.846
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr15_+_63050761
|
2.846
|
|
TLN2
|
talin 2
|
|
chr9_+_17578952
|
2.831
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr15_+_43885251
|
2.792
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr18_-_43678251
|
2.791
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr18_-_43678195
|
2.759
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr3_-_52488030
|
2.736
|
NM_003280
|
TNNC1
|
troponin C type 1 (slow)
|
|
chr12_+_109577201
|
2.717
|
NM_001093
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr15_-_75230354
|
2.713
|
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr9_-_138853146
|
2.712
|
NM_016172
|
UBAC1
|
UBA domain containing 1
|
|
chr19_-_4535202
|
2.680
|
NM_001013706
|
PLIN5
|
perilipin 5
|
|
chr6_-_33714681
|
2.674
|
NM_001142883
NM_054111
|
IP6K3
|
inositol hexakisphosphate kinase 3
|
|
chr1_-_116311330
|
2.672
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr11_-_47470687
|
2.672
|
NM_005055
NM_032645
|
RAPSN
|
receptor-associated protein of the synapse
|
|
chr1_-_17380486
|
2.668
|
NM_003000
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip)
|
|
chr1_-_116311161
|
2.638
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr4_-_111563278
|
2.629
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr11_-_73720281
|
2.610
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr18_-_43678233
|
2.606
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr12_-_96184346
|
2.601
|
|
NTN4
|
netrin 4
|
|
chr7_-_10979683
|
2.598
|
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr2_-_152590983
|
2.593
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr9_-_97356007
|
2.587
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr9_+_139846767
|
2.571
|
NM_178536
|
LCN12
|
lipocalin 12
|
|
chr4_+_72204769
|
2.543
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_64088886
|
2.539
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr16_+_30386097
|
2.525
|
NM_013292
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr2_+_103378489
|
2.485
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr16_+_72097124
|
2.475
|
NM_020995
|
HPR
|
haptoglobin-related protein
|
|
chr10_-_74714479
|
2.465
|
NM_032562
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr15_+_78441737
|
2.450
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr13_+_29598746
|
2.418
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr14_+_21498344
|
2.416
|
NM_173846
|
TPPP2
|
tubulin polymerization-promoting protein family member 2
|
|
chr8_+_145149985
|
2.403
|
|
CYC1
|
cytochrome c-1
|
|
chr4_-_177190258
|
2.399
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr5_+_53751430
|
2.390
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr19_+_35629727
|
2.381
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr1_-_151804225
|
2.369
|
NM_005060
|
RORC
|
RAR-related orphan receptor C
|
|
chr8_-_107782417
|
2.361
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr1_-_216978708
|
2.359
|
NM_001243514
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_+_68151092
|
2.350
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr9_-_138853007
|
2.345
|
|
UBAC1
|
UBA domain containing 1
|
|
chr11_+_34938112
|
2.338
|
|
PDHX
|
pyruvate dehydrogenase complex, component X
|
|
chr11_+_111896076
|
2.332
|
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr16_-_31439650
|
2.332
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr1_-_217113007
|
2.327
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr11_+_116700620
|
2.325
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr11_-_66139064
|
2.323
|
NM_001532
|
SLC29A2
|
solute carrier family 29 (nucleoside transporters), member 2
|
|
chr18_-_3219846
|
2.318
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr6_-_24489738
|
2.318
|
NM_177483
NM_001503
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1
|
|
chr18_-_43678257
|
2.309
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr3_+_158991035
|
2.306
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr20_-_33999760
|
2.303
|
|
UQCC
|
ubiquinol-cytochrome c reductase complex chaperone
|
|
chr1_-_27240430
|
2.292
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr18_-_44336991
|
2.258
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chrX_+_70521627
|
2.254
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr10_+_123872440
|
2.248
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_-_107582733
|
2.248
|
NM_003063
|
SLN
|
sarcolipin
|
|
chr5_-_150727088
|
2.229
|
NM_181776
|
SLC36A2
|
solute carrier family 36 (proton/amino acid symporter), member 2
|
|
chr19_+_33166312
|
2.211
|
NM_207391
|
RGS9BP
|
regulator of G protein signaling 9 binding protein
|
|
chrX_+_152954965
|
2.207
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr1_+_44412640
|
2.187
|
|
IPO13
|
importin 13
|
|
chr2_+_26467615
|
2.170
|
NM_000183
|
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit
|
|
chr17_+_42836546
|
2.158
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chr15_-_75230386
|
2.154
|
NM_004255
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr7_-_10979752
|
2.152
|
NM_002489
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
|
|
chr12_-_57039785
|
2.147
|
NM_001686
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr7_+_37960162
|
2.142
|
NM_001242946
NM_017549
|
EPDR1
|
ependymin related protein 1 (zebrafish)
|
|
chr7_+_123249076
|
2.135
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr8_-_124553253
|
2.126
|
|
FBXO32
|
F-box protein 32
|
|
chr12_-_57039736
|
2.124
|
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide
|
|
chr1_+_44412582
|
2.120
|
|
IPO13
|
importin 13
|
|
chr9_+_74764394
|
2.102
|
|
GDA
|
guanine deaminase
|
|
chr1_+_16348365
|
2.097
|
NM_001042704
NM_004070
|
CLCNKA
CLCNKB
|
chloride channel Ka
chloride channel Kb
|
|
chr20_+_17207680
|
2.095
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr12_-_96184510
|
2.088
|
NM_021229
|
NTN4
|
netrin 4
|
|
chr15_+_78441678
|
2.085
|
NM_005530
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr6_+_146348781
|
2.084
|
NM_000838
NM_001114329
|
GRM1
|
glutamate receptor, metabotropic 1
|
|
chr18_-_43678273
|
2.075
|
NM_004046
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr15_+_78441733
|
2.069
|
|
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr12_-_111358353
|
2.059
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr4_+_71494460
|
2.058
|
NM_031889
|
ENAM
|
enamelin
|
|
chrX_-_21676416
|
2.047
|
NM_153270
|
KLHL34
|
kelch-like 34 (Drosophila)
|
|
chr2_-_219696511
|
2.041
|
NM_017431
|
PRKAG3
|
protein kinase, AMP-activated, gamma 3 non-catalytic subunit
|
|
chr6_+_158734902
|
2.040
|
|
|
|
|
chr15_-_42783293
|
2.037
|
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chrX_-_129299680
|
2.026
|
|
AIFM1
|
apoptosis-inducing factor, mitochondrion-associated, 1
|
|
chr7_-_99381703
|
1.986
|
NM_001202855
NM_017460
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4
|
|
chr17_+_27370155
|
1.986
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr8_+_145729464
|
1.983
|
NM_005309
|
GPT
|
glutamic-pyruvate transaminase (alanine aminotransferase)
|
|
chr11_+_22696318
|
1.979
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chrX_+_135230736
|
1.974
|
NM_001159700
|
FHL1
|
four and a half LIM domains 1
|
|
chr16_+_31225341
|
1.971
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr12_-_81991959
|
1.971
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr15_+_52311432
|
1.968
|
|
MAPK6
|
mitogen-activated protein kinase 6
|
|
chr11_-_118550354
|
1.961
|
NM_007180
|
TREH
|
trehalase (brush-border membrane glycoprotein)
|
|
chr22_-_24110054
|
1.959
|
NM_213720
|
CHCHD10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr10_-_74714563
|
1.957
|
|
PLA2G12B
|
phospholipase A2, group XIIB
|
|
chr4_-_140216951
|
1.942
|
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa
|
|
chr3_-_38691118
|
1.938
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr5_-_133340380
|
1.933
|
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr2_+_26467809
|
1.933
|
|
HADHB
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit
|
|
chr11_+_20044362
|
1.933
|
|
NAV2
|
neuron navigator 2
|
|
chr5_-_133340389
|
1.931
|
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr1_+_54359859
|
1.928
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr12_-_122296619
|
1.923
|
NM_002150
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase
|
|
chr9_+_95947211
|
1.911
|
NM_006648
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chr5_-_133340358
|
1.901
|
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr2_-_21231406
|
1.900
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr4_-_8129378
|
1.867
|
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr1_-_46685962
|
1.866
|
NM_001243766
|
POMGNT1
|
protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase
|
|
chr20_+_57875647
|
1.860
|
|
EDN3
|
endothelin 3
|
|
chr19_+_13134782
|
1.856
|
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr11_-_19223516
|
1.853
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr5_-_133340427
|
1.850
|
|
VDAC1
|
voltage-dependent anion channel 1
|
|
chr17_-_27949814
|
1.841
|
|
CORO6
|
coronin 6
|
|
chr3_-_48647085
|
1.838
|
NM_003365
|
UQCRC1
|
ubiquinol-cytochrome c reductase core protein I
|
|
chr13_+_76334796
|
1.837
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr3_-_46718898
|
1.831
|
NM_182775
|
ALS2CL
|
ALS2 C-terminal like
|