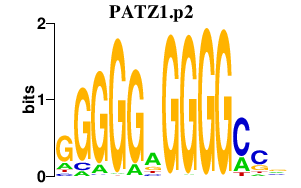
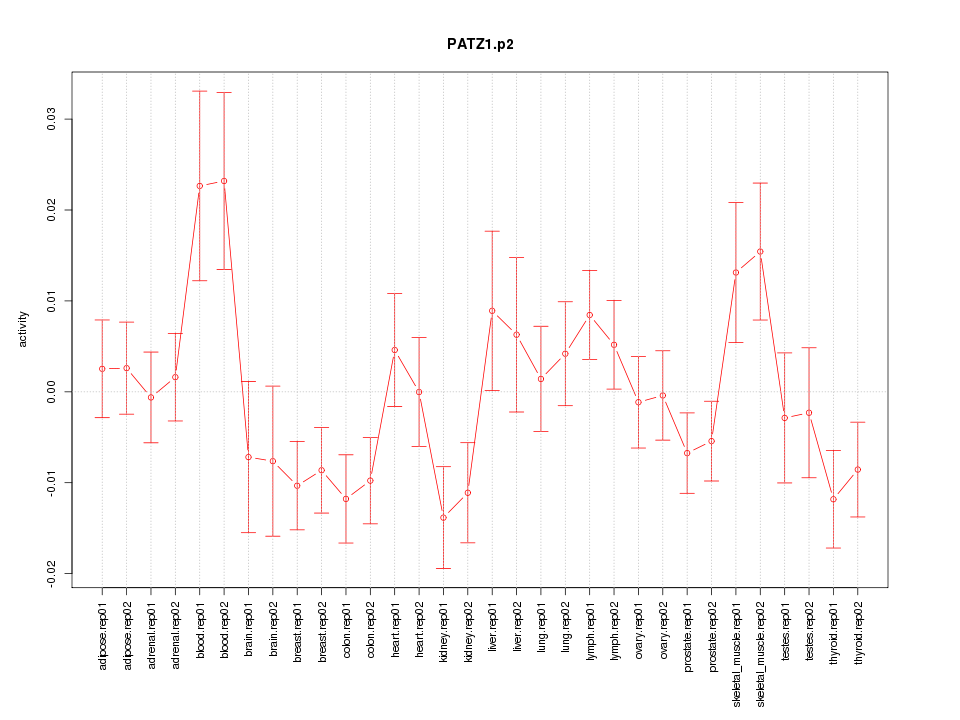
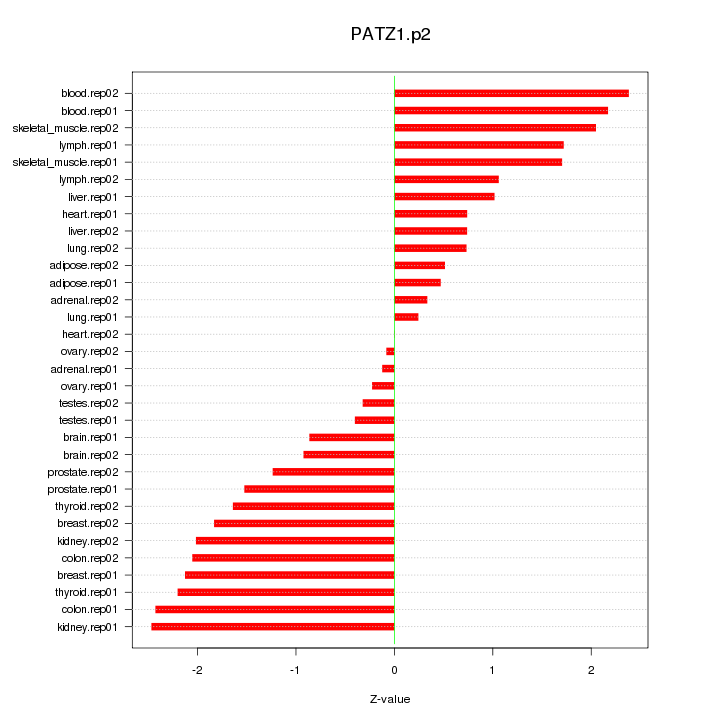
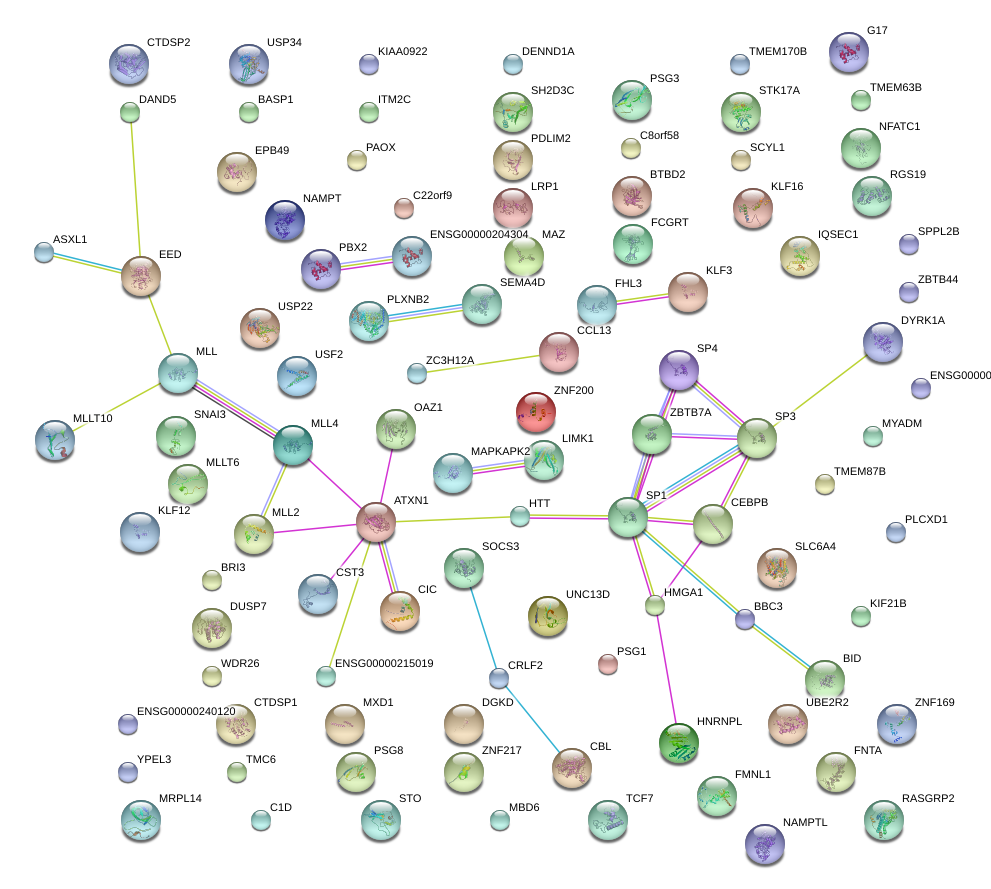
|
chr19_-_2015628
|
4.927
|
NM_017797
|
BTBD2
|
BTB (POZ) domain containing 2
|
|
chr18_+_77155714
|
4.110
|
NM_006162
NM_172388
NM_172390
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr20_-_62711160
|
3.586
|
NM_001039467
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr4_+_154387450
|
3.465
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr18_+_77155966
|
3.460
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr18_+_77155943
|
3.345
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr18_+_77155927
|
3.295
|
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr20_-_62710787
|
3.104
|
NM_005873
|
RGS19
|
regulator of G-protein signaling 19
|
|
chr16_-_3285359
|
3.050
|
NM_001145448
NM_198088
NM_001145446
NM_198087
|
ZNF200
|
zinc finger protein 200
|
|
chr16_-_88752879
|
3.038
|
NM_178310
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr12_+_57522429
|
2.983
|
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr16_-_88752788
|
2.957
|
|
SNAI3
|
snail homolog 3 (Drosophila)
|
|
chr20_-_52210780
|
2.940
|
|
ZNF217
|
zinc finger protein 217
|
|
chr7_+_21467572
|
2.665
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr8_+_22457098
|
2.606
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr6_-_44095177
|
2.494
|
NM_032111
|
MRPL14
|
mitochondrial ribosomal protein L14
|
|
chr2_-_174829108
|
2.384
|
|
SP3
|
Sp3 transcription factor
|
|
chr9_+_33817213
|
2.374
|
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr12_+_53774422
|
2.372
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr17_-_73840422
|
2.297
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr16_-_30107492
|
2.275
|
NM_031477
NM_001145524
|
YPEL3
|
yippee-like 3 (Drosophila)
|
|
chr12_+_53773924
|
2.266
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr17_+_36861857
|
2.249
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_219264719
|
2.217
|
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr12_-_58240746
|
2.166
|
NM_005730
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr2_+_112812799
|
2.163
|
NM_032824
|
TMEM87B
|
transmembrane protein 87B
|
|
chr6_+_34205369
|
2.155
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr11_-_64512265
|
2.145
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr11_+_119077021
|
2.141
|
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr1_-_224621714
|
2.104
|
|
WDR26
|
WD repeat domain 26
|
|
chr6_+_34205029
|
2.078
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr19_-_47734215
|
2.068
|
NM_014417
|
BBC3
|
BCL2 binding component 3
|
|
chr19_+_42788605
|
2.055
|
NM_015125
|
CIC
|
capicua homolog (Drosophila)
|
|
chr17_+_36861398
|
2.024
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr7_+_73498106
|
2.024
|
NM_002314
|
LIMK1
|
LIM domain kinase 1
|
|
chr2_+_70142303
|
1.978
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr22_-_50746000
|
1.968
|
NM_012401
|
PLXNB2
|
plexin B2
|
|
chr7_+_97910978
|
1.956
|
NM_001159491
NM_015379
|
BRI3
|
brain protein I3
|
|
chr5_+_133450353
|
1.934
|
NM_003202
|
TCF7
|
transcription factor 7 (T-cell specific, HMG-box)
|
|
chr2_+_234263153
|
1.933
|
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr12_-_58240217
|
1.924
|
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr6_+_44095357
|
1.921
|
NM_018426
|
TMEM63B
|
transmembrane protein 63B
|
|
chr10_+_135192694
|
1.912
|
NM_152911
NM_207127
NM_207128
|
PAOX
|
polyamine oxidase (exo-N4-amino)
|
|
chr3_-_52090090
|
1.912
|
NM_001947
|
DUSP7
|
dual specificity phosphatase 7
|
|
chr17_-_20946088
|
1.906
|
NM_015276
|
USP22
|
ubiquitin specific peptidase 22
|
|
chr1_+_37940129
|
1.881
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr9_-_130517552
|
1.869
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr19_+_35759967
|
1.867
|
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr7_+_73498149
|
1.857
|
|
LIMK1
|
LIM domain kinase 1
|
|
chr12_+_57522247
|
1.847
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chrX_-_1331526
|
1.839
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr13_-_74708361
|
1.838
|
|
KLF12
|
Kruppel-like factor 12
|
|
chrY_-_1281526
|
1.835
|
NM_001012288
NM_022148
|
CRLF2
|
cytokine receptor-like factor 2
|
|
chr6_+_11538486
|
1.834
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr5_+_176560627
|
1.830
|
NM_022455
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr4_+_3076236
|
1.808
|
NM_002111
|
HTT
|
huntingtin
|
|
chr16_+_29817810
|
1.785
|
NM_001042539
NM_002383
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr12_+_57916607
|
1.781
|
NM_052897
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr1_+_37940180
|
1.777
|
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr2_+_112812900
|
1.744
|
|
TMEM87B
|
transmembrane protein 87B
|
|
chr6_-_44095153
|
1.740
|
|
MRPL14
|
mitochondrial ribosomal protein L14
|
|
chr19_-_1863425
|
1.728
|
|
KLF16
|
Kruppel-like factor 16
|
|
chr19_-_913194
|
1.723
|
|
R3HDM4
|
R3H domain containing 4
|
|
chr19_+_50016500
|
1.722
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr20_+_48807460
|
1.712
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr3_-_13008959
|
1.712
|
NM_014869
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr1_-_224622000
|
1.709
|
NM_001115113
NM_025160
|
WDR26
|
WD repeat domain 26
|
|
chr22_-_18257160
|
1.690
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr1_-_38471170
|
1.689
|
NM_001243878
NM_004468
|
FHL3
|
four and a half LIM domains 3
|
|
chr6_-_32157511
|
1.686
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr11_+_119076974
|
1.679
|
NM_005188
|
CBL
|
Cas-Br-M (murine) ecotropic retroviral transforming sequence
|
|
chr20_+_30946786
|
1.673
|
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr7_+_43622638
|
1.653
|
NM_004760
|
STK17A
|
serine/threonine kinase 17a
|
|
chr19_-_4066739
|
1.648
|
NM_015898
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr2_-_61697846
|
1.646
|
NM_014709
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr1_+_206858308
|
1.645
|
NM_004759
NM_032960
|
MAPKAPK2
|
mitogen-activated protein kinase-activated protein kinase 2
|
|
chr17_-_73840495
|
1.634
|
|
UNC13D
|
unc-13 homolog D (C. elegans)
|
|
chr17_-_76356153
|
1.618
|
NM_003955
|
SOCS3
|
suppressor of cytokine signaling 3
|
|
chr17_-_76124767
|
1.615
|
|
TMC6
|
transmembrane channel-like 6
|
|
chr13_-_74707913
|
1.595
|
NM_007249
|
KLF12
|
Kruppel-like factor 12
|
|
chr5_+_17217474
|
1.589
|
NM_006317
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chrX_+_198060
|
1.589
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr1_-_200992510
|
1.584
|
|
KIF21B
|
kinesin family member 21B
|
|
chr9_-_92112887
|
1.577
|
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D
|
|
chr11_+_118307072
|
1.575
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr22_-_18256748
|
1.564
|
NM_001244572
NM_197966
|
BID
|
BH3 interacting domain death agonist
|
|
chr22_-_45636584
|
1.560
|
NM_001009880
|
KIAA0930
|
KIAA0930
|
|
chr6_-_16761600
|
1.557
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr19_+_54371150
|
1.535
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_+_37940074
|
1.532
|
NM_025079
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr11_-_130184459
|
1.525
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr21_+_38739193
|
1.523
|
|
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A
|
|
chr9_+_33817168
|
1.522
|
NM_017811
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr19_-_39342957
|
1.519
|
NM_001005335
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L
|
|
chr4_+_38665587
|
1.515
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr17_+_43299277
|
1.502
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr11_+_65292576
|
1.501
|
|
SCYL1
|
SCY1-like 1 (S. cerevisiae)
|
|
chr19_+_2269515
|
1.499
|
NM_004152
|
OAZ1
SPPL2B
|
ornithine decarboxylase antizyme 1
signal peptide peptidase-like 2B
|
|
chr22_-_18257238
|
1.499
|
NM_001196
NM_001244569
NM_001244570
NM_197967
NM_001244567
|
BID
|
BH3 interacting domain death agonist
|
|
chr19_+_50016435
|
1.498
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_-_105925329
|
1.480
|
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr19_+_36209125
|
1.476
|
|
MLL4
|
myeloid/lymphoid or mixed-lineage leukemia 4
|
|
chr9_-_126692184
|
1.473
|
NM_020946
NM_024820
|
DENND1A
|
DENN/MADD domain containing 1A
|
|
chr20_-_23618382
|
1.467
|
NM_000099
|
CST3
|
cystatin C
|
|
chr12_-_58240493
|
1.460
|
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr6_+_34204928
|
1.456
|
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr7_-_100487273
|
1.454
|
NM_001015072
|
UFSP1
|
UFM1-specific peptidase 1 (non-functional)
|
|
chr19_+_50016490
|
1.449
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr9_+_137218313
|
1.448
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr2_+_70142236
|
1.446
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr6_-_91006460
|
1.444
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr11_+_64949149
|
1.441
|
NM_001198869
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr7_+_97911023
|
1.433
|
|
BRI3
|
brain protein I3
|
|
chr1_-_36948423
|
1.432
|
|
CSF3R
|
colony stimulating factor 3 receptor (granulocyte)
|
|
chr5_+_172261220
|
1.428
|
NM_001031711
|
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1
|
|
chr20_-_4804067
|
1.428
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr19_+_35759880
|
1.423
|
NM_003367
NM_207291
|
USF2
|
upstream transcription factor 2, c-fos interacting
|
|
chr8_+_1711869
|
1.420
|
NM_018941
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr11_-_3862137
|
1.414
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr7_-_100239136
|
1.411
|
NM_003227
|
TFR2
|
transferrin receptor 2
|
|
chr8_+_1712011
|
1.410
|
|
CLN8
|
ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation)
|
|
chr6_-_30655078
|
1.409
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr1_-_19812004
|
1.408
|
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta
|
|
chr6_-_91006580
|
1.399
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_-_43453146
|
1.399
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr20_+_34894190
|
1.398
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr11_+_64948685
|
1.397
|
NM_001198868
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr7_-_105925410
|
1.391
|
NM_005746
|
NAMPT
|
nicotinamide phosphoribosyltransferase
|
|
chr1_+_226250413
|
1.387
|
|
H3F3A
|
H3 histone, family 3A
|
|
chr2_-_61697899
|
1.387
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr1_+_206680863
|
1.385
|
NM_182663
NM_182664
|
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr2_+_70142152
|
1.379
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr8_+_144635556
|
1.379
|
NM_001166237
|
GSDMD
|
gasdermin D
|
|
chr2_+_234263141
|
1.375
|
NM_152879
|
DGKD
|
diacylglycerol kinase, delta 130kDa
|
|
chr11_-_130184314
|
1.373
|
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr9_-_110251754
|
1.371
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr2_-_16847070
|
1.371
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr22_-_18257221
|
1.369
|
|
BID
|
BH3 interacting domain death agonist
|
|
chr19_+_54371125
|
1.357
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr9_+_33817590
|
1.356
|
|
UBE2R2
|
ubiquitin-conjugating enzyme E2R 2
|
|
chr16_+_90015827
|
1.356
|
NM_001242819
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr12_-_116714910
|
1.351
|
NM_015335
|
MED13L
|
mediator complex subunit 13-like
|
|
chr9_+_129622935
|
1.351
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr1_-_19812065
|
1.347
|
NM_001206540
NM_004930
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta
|
|
chr2_-_43453724
|
1.339
|
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr22_-_26986071
|
1.338
|
NM_003595
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr12_-_58240472
|
1.335
|
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr18_+_77160243
|
1.331
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr17_+_38278789
|
1.325
|
NM_001012241
|
MSL1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chr11_-_3862183
|
1.324
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr21_+_30671113
|
1.315
|
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1
|
|
chr16_+_90015138
|
1.314
|
NM_001242816
NM_001242817
NM_001242818
NM_001242820
NM_001242821
NM_001242822
NM_017702
NM_207514
|
DEF8
|
differentially expressed in FDCP 8 homolog (mouse)
|
|
chr5_+_17217781
|
1.313
|
|
BASP1
|
brain abundant, membrane attached signal protein 1
|
|
chr3_+_184032849
|
1.312
|
NM_182917
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr11_+_64949413
|
1.311
|
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr20_+_48807119
|
1.304
|
NM_005194
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr11_-_64510461
|
1.302
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr11_+_64949279
|
1.302
|
NM_005186
|
CAPN1
|
calpain 1, (mu/I) large subunit
|
|
chr17_+_55333843
|
1.298
|
NM_138962
|
MSI2
|
musashi homolog 2 (Drosophila)
|
|
chr19_+_30156326
|
1.295
|
NM_024310
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr1_-_19811951
|
1.294
|
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta
|
|
chr2_+_238600780
|
1.292
|
NM_001137551
NM_001137552
NM_001137553
NM_004735
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr22_-_26986065
|
1.289
|
|
TPST2
|
tyrosylprotein sulfotransferase 2
|
|
chr19_+_54372659
|
1.286
|
NM_001020821
NM_001020818
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_2269526
|
1.278
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr6_-_30654978
|
1.275
|
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr19_+_49259343
|
1.275
|
NM_019113
|
FGF21
|
fibroblast growth factor 21
|
|
chr18_+_11981421
|
1.268
|
NM_014214
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr17_-_43319100
|
1.266
|
|
|
|
|
chr12_-_58240446
|
1.260
|
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr6_+_37137882
|
1.257
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr1_+_154975041
|
1.255
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr17_-_74497406
|
1.255
|
NM_024599
|
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila)
|
|
chr9_-_35115844
|
1.252
|
NM_025182
|
FAM214B
|
family with sequence similarity 214, member B
|
|
chr10_+_104155431
|
1.250
|
NM_002502
|
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100)
|
|
chr1_-_200992824
|
1.249
|
NM_001252100
NM_001252102
NM_001252103
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr16_+_68119211
|
1.246
|
NM_004555
NM_173163
NM_173165
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr19_-_10628606
|
1.243
|
NM_001166215
|
S1PR5
|
sphingosine-1-phosphate receptor 5
|
|
chr8_-_67525426
|
1.240
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr12_-_58132028
|
1.237
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr6_+_29691203
|
1.234
|
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr19_-_913224
|
1.224
|
NM_138774
|
R3HDM4
|
R3H domain containing 4
|
|
chr4_-_1166547
|
1.224
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr2_+_238600878
|
1.223
|
|
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1
|
|
chr18_+_11981552
|
1.221
|
|
IMPA2
|
inositol(myo)-1(or 4)-monophosphatase 2
|
|
chr7_+_143078358
|
1.217
|
NM_001010972
NM_003461
|
ZYX
|
zyxin
|
|
chr19_-_2038937
|
1.215
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr17_+_76164624
|
1.215
|
NM_004710
|
SYNGR2
|
synaptogyrin 2
|
|
chr5_+_176560882
|
1.213
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr5_+_137673956
|
1.213
|
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr20_+_48807365
|
1.213
|
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr16_+_29817426
|
1.212
|
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chr19_+_54371170
|
1.202
|
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr19_+_2269529
|
1.201
|
|
OAZ1
|
ornithine decarboxylase antizyme 1
|
|
chr4_-_1166364
|
1.201
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr19_-_4065467
|
1.200
|
|
ZBTB7A
|
zinc finger and BTB domain containing 7A
|
|
chr6_-_31324898
|
1.199
|
|
HLA-B
|
major histocompatibility complex, class I, B
|
|
chr1_-_25256767
|
1.196
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_-_64510339
|
1.194
|
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr7_+_73498318
|
1.192
|
|
LIMK1
|
LIM domain kinase 1
|