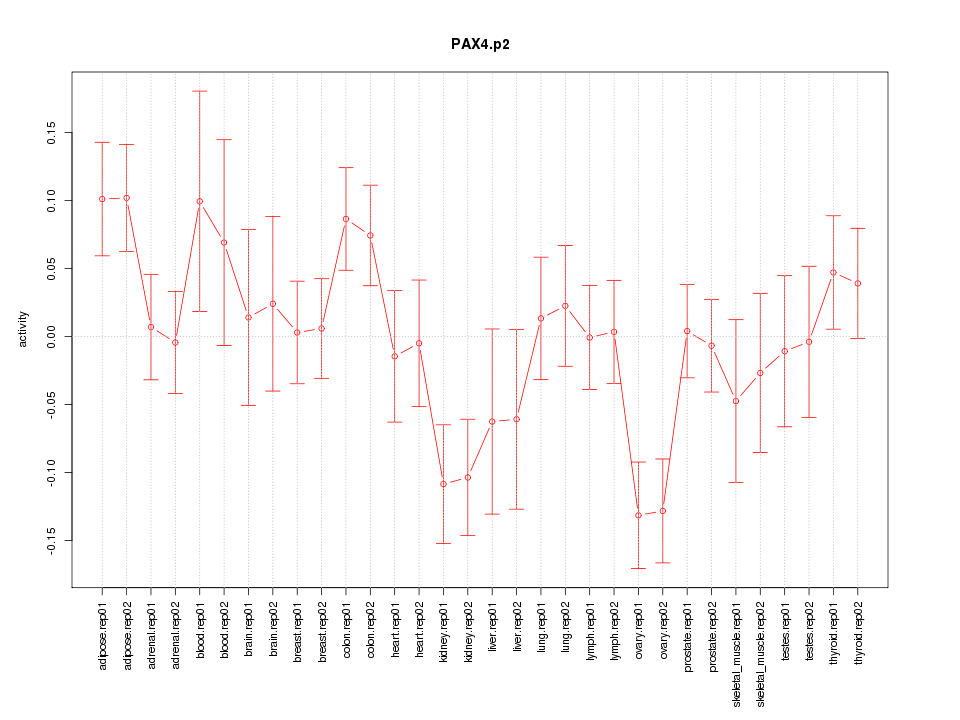
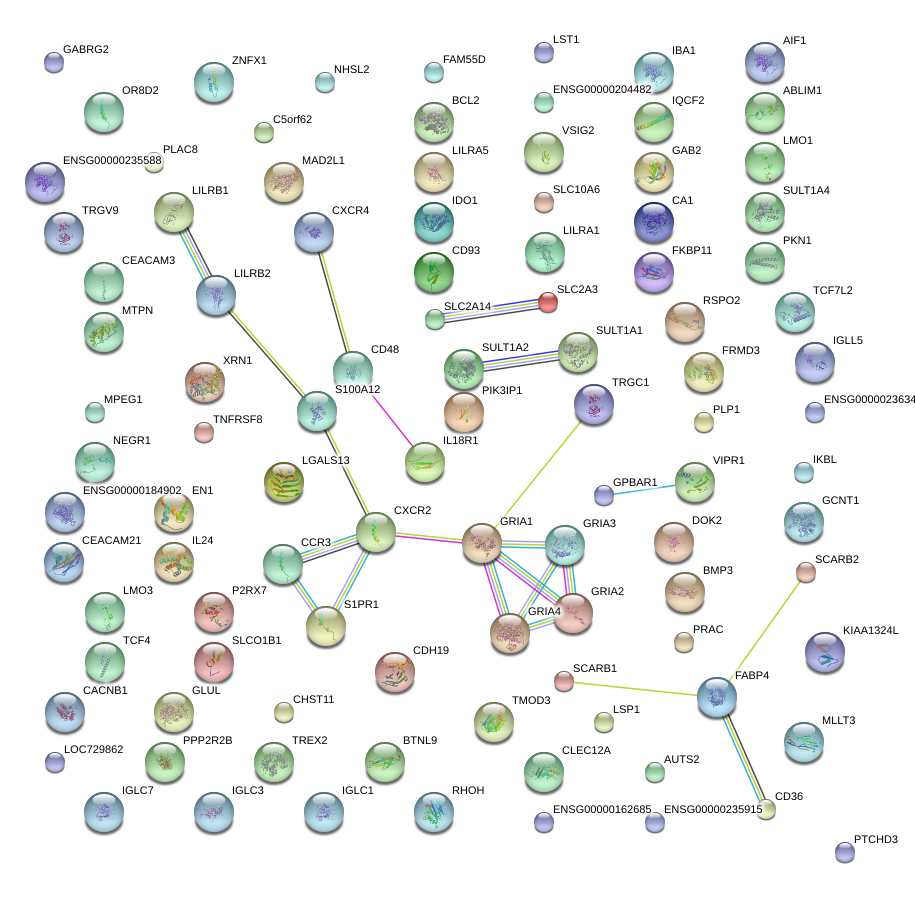
|
chr7_+_80267972
|
7.357
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr7_+_80267922
|
7.259
|
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr7_+_80267782
|
6.051
|
NM_000072
NM_001001548
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr1_+_101702561
|
4.037
|
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr1_-_153348057
|
3.711
|
NM_005621
|
S100A12
|
S100 calcium binding protein A12
|
|
chr1_+_101702304
|
2.875
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr19_-_54784922
|
2.192
|
NM_001080978
NM_005874
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
|
|
chr12_-_16759531
|
1.959
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr20_-_23062786
|
1.936
|
|
CD93
|
CD93 molecule
|
|
chr19_+_42300521
|
1.884
|
NM_001815
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3
|
|
chr19_-_54824343
|
1.853
|
NM_021250
NM_181879
NM_181985
NM_181986
|
LILRA5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5
|
|
chr10_-_27703178
|
1.781
|
NM_001034842
|
PTCHD3
|
patched domain containing 3
|
|
chrX_+_71130619
|
1.766
|
NM_001013627
|
NHSL2
|
NHS-like 2
|
|
chr9_-_20622476
|
1.736
|
NM_004529
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr8_-_86290333
|
1.646
|
NM_001128829
NM_001128830
NM_001128831
NM_001738
|
CA1
|
carbonic anhydrase I
|
|
chr9_-_20622433
|
1.640
|
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr2_+_219125737
|
1.633
|
NM_001077191
NM_001077194
NM_170699
|
GPBAR1
|
G protein-coupled bile acid receptor 1
|
|
chr8_-_82395401
|
1.517
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_-_160681552
|
1.516
|
NM_001778
|
CD48
|
CD48 molecule
|
|
chr3_+_42530790
|
1.485
|
NM_001251882
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|
|
chr8_-_21771111
|
1.469
|
NM_003974
|
DOK2
|
docking protein 2, 56kDa
|
|
chr19_+_55141907
|
1.382
|
NM_001081638
NM_001081639
NM_001081637
|
LILRB1
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1
|
|
chr11_-_124622133
|
1.377
|
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr4_-_84030995
|
1.374
|
NM_001130716
|
PLAC8
|
placenta-specific 8
|
|
chr6_+_31554475
|
1.364
|
NM_205837
|
LST1
|
leukocyte specific transcript 1
|
|
chr9_+_79115548
|
1.359
|
NM_001097636
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr22_-_31688329
|
1.334
|
NM_001135911
NM_052880
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr17_-_46799879
|
1.310
|
NM_032391
|
PRAC
|
prostate cancer susceptibility candidate
|
|
chr11_+_1874199
|
1.310
|
NM_002339
|
LSP1
|
lymphocyte-specific protein 1
|
|
chr8_-_13133956
|
1.307
|
|
|
|
|
chr7_-_135613197
|
1.287
|
|
LUZP6
|
leucine zipper protein 6
|
|
chr12_-_16760935
|
1.273
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr9_-_85882093
|
1.249
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr1_+_12123433
|
1.249
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr18_-_64271215
|
1.245
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr2_-_136873740
|
1.236
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr11_-_58980321
|
1.229
|
NM_001039396
|
MPEG1
|
macrophage expressed 1
|
|
chr7_-_86595203
|
1.211
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr6_+_31553955
|
1.210
|
NM_205838
NM_205839
|
LST1
|
leukocyte specific transcript 1
|
|
chr5_+_150157507
|
1.195
|
NM_032947
|
C5orf62
|
chromosome 5 open reading frame 62
|
|
chr16_-_28608367
|
1.195
|
NM_001054
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
|
|
chrX_+_103031433
|
1.188
|
NM_001128834
|
PLP1
|
proteolipid protein 1
|
|
chr5_+_152870083
|
1.181
|
NM_000827
NM_001114183
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1
|
|
chrX_+_103031753
|
1.174
|
NM_000533
NM_199478
|
PLP1
|
proteolipid protein 1
|
|
chr22_+_23229959
|
1.157
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr3_+_46283871
|
1.156
|
NM_001164680
NM_001837
NM_178328
NM_178329
|
CCR3
|
chemokine (C-C motif) receptor 3
|
|
chr2_-_119605758
|
1.149
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr11_-_78052925
|
1.128
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr18_-_53089722
|
1.117
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr12_+_105153258
|
1.117
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr18_-_53255734
|
1.112
|
NM_001083962
NM_001243230
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr18_-_60795878
|
1.080
|
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr2_+_102979096
|
1.075
|
NM_003855
|
IL18R1
|
interleukin 18 receptor 1
|
|
chr12_-_16759430
|
1.066
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_+_161494647
|
1.042
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr7_-_38313232
|
1.031
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr2_+_218990735
|
1.024
|
NM_001557
|
CXCR2
|
chemokine (C-X-C motif) receptor 2
|
|
chr1_-_72748147
|
1.023
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr5_+_180467224
|
1.018
|
NM_152547
|
BTNL9
|
butyrophilin-like 9
|
|
chr1_-_72748491
|
0.999
|
|
|
|
|
chr6_+_29894105
|
0.994
|
|
|
|
|
chr6_+_31582978
|
0.988
|
NM_004847
NM_001623
|
AIF1
|
allograft inflammatory factor 1
|
|
chr8_-_109095836
|
0.988
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chr4_+_81952118
|
0.986
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr12_+_121570630
|
0.968
|
NM_002562
|
P2RX7
|
purinergic receptor P2X, ligand-gated ion channel, 7
|
|
chr3_-_142166862
|
0.966
|
|
XRN1
|
5'-3' exoribonuclease 1
|
|
chr19_-_4916814
|
0.950
|
|
|
|
|
chr1_+_207070787
|
0.939
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr1_-_182357757
|
0.916
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_+_51895644
|
0.916
|
NM_203424
|
IQCF2
|
IQ motif containing F2
|
|
chr19_+_55105046
|
0.899
|
NM_006863
|
LILRA1
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1
|
|
chr4_+_40198526
|
0.885
|
NM_004310
|
RHOH
|
ras homolog gene family, member H
|
|
chr12_-_8088629
|
0.875
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr15_+_52201590
|
0.872
|
|
TMOD3
|
tropomodulin 3 (ubiquitous)
|
|
chr11_-_114466471
|
0.855
|
NM_001077639
NM_017678
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr8_+_39771327
|
0.828
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr10_-_116286570
|
0.824
|
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr4_-_87770267
|
0.820
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr5_-_146460991
|
0.815
|
NM_181677
NM_181678
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr12_-_16758839
|
0.798
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_+_95326397
|
0.797
|
NM_001195755
NM_181745
|
O3FAR1
|
omega-3 fatty acid receptor 1
|
|
chr19_+_14544216
|
0.787
|
|
PKN1
|
protein kinase N1
|
|
chr12_+_10124007
|
0.786
|
NM_138337
NM_201623
|
CLEC12A
CLEC12B
|
C-type lectin domain family 12, member A
C-type lectin domain family 12, member B
|
|
chr20_-_47874147
|
0.768
|
|
ZNFX1
|
zinc finger, NFX1-type containing 1
|
|
chr4_-_120987890
|
0.767
|
NM_002358
|
MAD2L1
|
MAD2 mitotic arrest deficient-like 1 (yeast)
|
|
chr1_-_182357871
|
0.767
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr7_+_69064580
|
0.764
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr14_-_69259654
|
0.751
|
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr11_+_63273555
|
0.751
|
|
LGALS12
|
lectin, galactoside-binding, soluble, 12
|
|
chr3_-_21792815
|
0.745
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr2_-_96987618
|
0.744
|
|
|
|
|
chr5_+_139876504
|
0.733
|
|
ANKHD1
|
ankyrin repeat and KH domain containing 1
|
|
chr11_+_114549199
|
0.732
|
NM_182495
|
FAM55B
|
family with sequence similarity 55, member B
|
|
chr15_+_67418053
|
0.725
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_175395544
|
0.725
|
NM_032361
|
THOC3
|
THO complex 3
|
|
chr1_+_26644380
|
0.720
|
NM_001803
|
CD52
|
CD52 molecule
|
|
chr1_-_182360889
|
0.718
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_+_111260925
|
0.712
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr16_+_31366468
|
0.712
|
NM_000887
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit)
|
|
chr12_-_120687956
|
0.708
|
NM_025157
|
PXN
|
paxillin
|
|
chr16_-_84273303
|
0.705
|
NM_172347
|
KCNG4
|
potassium voltage-gated channel, subfamily G, member 4
|
|
chr5_-_58882274
|
0.696
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_-_151032082
|
0.694
|
NM_001038707
NM_020239
|
CDC42SE1
|
CDC42 small effector 1
|
|
chr7_-_23509972
|
0.686
|
NM_006547
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3
|
|
chr3_-_9921933
|
0.682
|
NM_001199551
NM_001199552
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr4_-_82126200
|
0.677
|
NM_006259
|
PRKG2
|
protein kinase, cGMP-dependent, type II
|
|
chr2_-_214015053
|
0.674
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chrX_+_155227245
|
0.666
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr19_+_10959105
|
0.664
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr18_-_60986508
|
0.663
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chrY_+_59330251
|
0.661
|
NM_002186
|
IL9R
|
interleukin 9 receptor
|
|
chr12_-_8693391
|
0.661
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr22_-_39640956
|
0.660
|
NM_002608
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr1_-_20445991
|
0.659
|
NM_012400
|
PLA2G2D
|
phospholipase A2, group IID
|
|
chr1_-_182360181
|
0.654
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr3_+_122296432
|
0.654
|
NM_001113523
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr6_+_32407618
|
0.653
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr6_+_29855349
|
0.649
|
|
HLA-H
|
major histocompatibility complex, class I, H (pseudogene)
|
|
chr12_+_104850986
|
0.648
|
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_+_116997185
|
0.645
|
NM_001085481
|
MAP1LC3B2
|
microtubule-associated protein 1 light chain 3 beta 2
|
|
chr4_-_176733413
|
0.642
|
|
GPM6A
|
glycoprotein M6A
|
|
chr3_+_119298522
|
0.640
|
NM_001125
|
ADPRH
|
ADP-ribosylarginine hydrolase
|
|
chr11_-_114466397
|
0.640
|
|
FAM55D
|
family with sequence similarity 55, member D
|
|
chr16_+_50731049
|
0.637
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr5_-_175394949
|
0.637
|
|
THOC3
|
THO complex 3
|
|
chr6_-_32557431
|
0.632
|
NM_001243965
NM_002124
|
HLA-DRB1
HLA-DRB4
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 1
major histocompatibility complex, class II, DR beta 4
major histocompatibility complex, class II, DR beta 5
|
|
chr16_+_71879858
|
0.632
|
|
|
|
|
chr12_-_95044205
|
0.625
|
NM_020698
|
TMCC3
|
transmembrane and coiled-coil domain family 3
|
|
chr3_-_71632741
|
0.618
|
NM_001244808
NM_001012505
NM_001244810
NM_032682
|
FOXP1
|
forkhead box P1
|
|
chr21_+_19289656
|
0.610
|
NM_001204175
NM_001204176
NM_001204177
NM_001204178
|
CHODL
|
chondrolectin
|
|
chr4_-_176733378
|
0.605
|
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_134233848
|
0.604
|
NM_001080538
|
AKR1B15
|
aldo-keto reductase family 1, member B15
|
|
chr17_-_1412995
|
0.599
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr22_-_17700265
|
0.598
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chr16_-_28621279
|
0.597
|
NM_001055
NM_177529
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
|
|
chr19_-_45996470
|
0.592
|
NM_206901
|
RTN2
|
reticulon 2
|
|
chrX_-_101410761
|
0.588
|
NM_001159560
NM_001012978
|
BEX5
|
brain expressed, X-linked 5
|
|
chr17_+_72322350
|
0.585
|
NM_153209
|
KIF19
|
kinesin family member 19
|
|
chr3_+_194406614
|
0.577
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr11_-_94964209
|
0.576
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr7_-_83278478
|
0.570
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr11_-_74109417
|
0.569
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr19_-_16250695
|
0.568
|
|
|
|
|
chr7_-_96654128
|
0.567
|
NM_005221
|
DLX5
|
distal-less homeobox 5
|
|
chr5_+_108083522
|
0.551
|
NM_005246
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr12_-_16759420
|
0.547
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_-_95748688
|
0.540
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr6_-_137494719
|
0.534
|
NM_052962
NM_181309
NM_181310
|
IL22RA2
|
interleukin 22 receptor, alpha 2
|
|
chr22_-_32651317
|
0.526
|
NM_014227
|
SLC5A4
|
solute carrier family 5 (low affinity glucose cotransporter), member 4
|
|
chr6_-_31560497
|
0.524
|
NM_001145466
NM_001145467
NM_147130
|
NCR3
|
natural cytotoxicity triggering receptor 3
|
|
chr5_-_142783059
|
0.523
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr7_+_72848108
|
0.522
|
NM_003508
|
FZD9
|
frizzled family receptor 9
|
|
chr11_-_124622034
|
0.521
|
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr6_-_149806025
|
0.517
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr20_-_52210780
|
0.514
|
|
ZNF217
|
zinc finger protein 217
|
|
chr11_+_60102294
|
0.503
|
NM_139249
|
MS4A6E
|
membrane-spanning 4-domains, subfamily A, member 6E
|
|
chr12_-_57644905
|
0.502
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr5_+_147258273
|
0.496
|
NM_054023
|
SCGB3A2
|
secretoglobin, family 3A, member 2
|
|
chr19_+_14544100
|
0.491
|
NM_002741
|
PKN1
|
protein kinase N1
|
|
chr4_-_176733901
|
0.488
|
|
GPM6A
|
glycoprotein M6A
|
|
chr7_+_18330044
|
0.488
|
|
HDAC9
|
histone deacetylase 9
|
|
chr18_-_60987212
|
0.486
|
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr18_-_53255361
|
0.486
|
|
TCF4
|
transcription factor 4
|
|
chr6_-_32805819
|
0.485
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr12_-_95945226
|
0.485
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr13_-_41593491
|
0.483
|
NM_172373
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr6_-_32806009
|
0.483
|
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr17_+_72363545
|
0.480
|
NM_181790
|
GPR142
|
G protein-coupled receptor 142
|
|
chr5_-_142783288
|
0.480
|
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr2_-_136875629
|
0.474
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr17_-_41738901
|
0.474
|
NM_004527
NM_013999
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr7_+_26191732
|
0.473
|
NM_004289
|
NFE2L3
|
nuclear factor (erythroid-derived 2)-like 3
|
|
chr9_-_27573444
|
0.471
|
NM_018325
|
C9orf72
|
chromosome 9 open reading frame 72
|
|
chr13_-_114898013
|
0.470
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr4_-_140060601
|
0.469
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chrX_+_2609224
|
0.468
|
NM_001122898
NM_002414
|
CD99
|
CD99 molecule
|
|
chr11_-_124622085
|
0.453
|
NM_014312
|
VSIG2
|
V-set and immunoglobulin domain containing 2
|
|
chr12_-_10562142
|
0.451
|
NM_013431
|
KLRC4-KLRK1
KLRC4
|
KLRC4-KLRK1 readthrough
killer cell lectin-like receptor subfamily C, member 4
|
|
chr1_+_26644458
|
0.450
|
|
CD52
|
CD52 molecule
|
|
chr17_+_43299277
|
0.450
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr13_-_114898057
|
0.450
|
|
RASA3
|
RAS p21 protein activator 3
|
|
chr3_+_69134057
|
0.448
|
NM_006407
|
ARL6IP5
|
ADP-ribosylation-like factor 6 interacting protein 5
|
|
chrX_-_19688991
|
0.436
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr9_-_91793375
|
0.431
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr9_-_123676849
|
0.430
|
NM_001190947
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr2_+_157291979
|
0.429
|
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
|
|
chr1_+_168545710
|
0.426
|
NM_002995
|
XCL1
|
chemokine (C motif) ligand 1
|
|
chr1_-_26699225
|
0.426
|
NM_001114759
NM_173574
|
ZNF683
|
zinc finger protein 683
|
|
chr12_+_10460416
|
0.421
|
NM_002262
NM_007334
|
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1
|
|
chr4_-_109089444
|
0.419
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr9_-_135282164
|
0.417
|
NM_001205296
NM_007344
|
TTF1
|
transcription termination factor, RNA polymerase I
|
|
chr6_+_29910246
|
0.416
|
NM_001242758
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr22_+_37447615
|
0.416
|
NM_024681
|
KCTD17
|
potassium channel tetramerisation domain containing 17
|
|
chrY_+_2559224
|
0.414
|
NM_001122898
NM_002414
|
CD99
|
CD99 molecule
|
|
chr2_-_87018802
|
0.414
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr14_-_92333856
|
0.406
|
NM_001128596
|
TC2N
|
tandem C2 domains, nuclear
|
|
chr2_+_122407564
|
0.405
|
|
LOC254128
|
uncharacterized LOC254128
|
|
chr22_+_36960325
|
0.403
|
|
|
|
|
chr10_+_114135013
|
0.402
|
NM_203380
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr1_+_25598883
|
0.402
|
NM_001127691
NM_016124
|
RHD
|
Rh blood group, D antigen
|