|
chr2_-_216241321
|
12.718
|
|
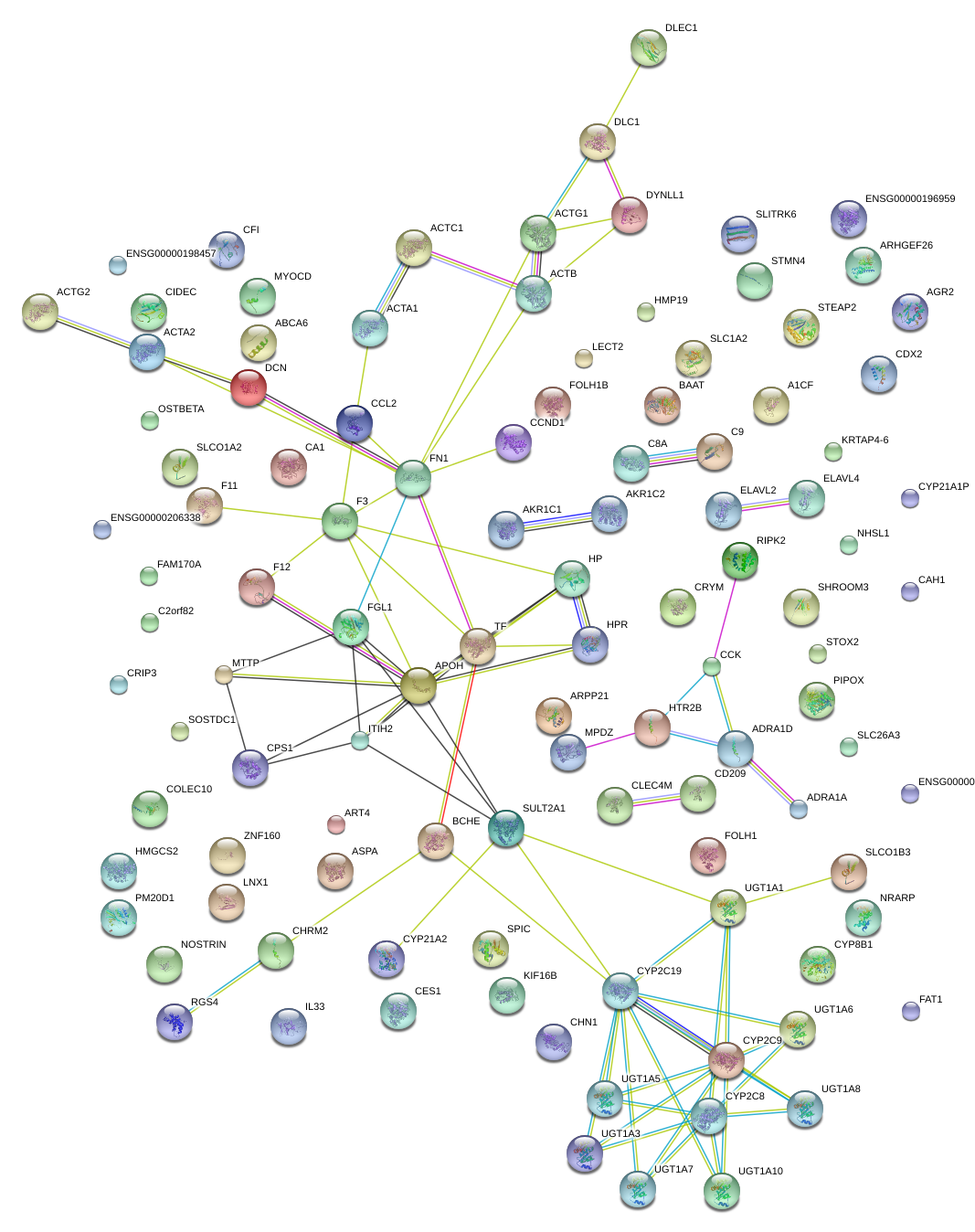
FN1
|
fibronectin 1
|
|
chr3_+_133494275
|
5.563
|
|
TF
|
transferrin
|
|
chr4_+_100495972
|
4.978
|
|
MTTP
|
microsomal triglyceride transfer protein
|
|
chr10_+_7745325
|
4.528
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr8_+_120079423
|
4.348
|
NM_006438
|
COLEC10
|
collectin sub-family member 10 (C-type lectin)
|
|
chr11_+_69455938
|
4.342
|
|
CCND1
|
cyclin D1
|
|
chr8_-_17752847
|
4.306
|
NM_147203
NM_201553
NM_004467
NM_201552
|
FGL1
|
fibrinogen-like 1
|
|
chr12_-_91573264
|
4.298
|
NM_133503
|
DCN
|
decorin
|
|
chr17_+_27370155
|
4.291
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr16_-_55867016
|
4.287
|
NM_001025194
NM_001025195
NM_001266
|
CES1
|
carboxylesterase 1
|
|
chr11_-_35441501
|
4.283
|
NM_001195728
NM_001252652
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr9_+_6215785
|
4.178
|
NM_001199640
NM_001199641
NM_033439
|
IL33
|
interleukin 33
|
|
chr10_+_7745229
|
4.064
|
NM_002216
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr10_+_7745284
|
3.959
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr11_-_49229994
|
3.918
|
NM_001014986
NM_001193471
NM_001193472
NM_001193473
NM_004476
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1
|
|
chr11_+_69455884
|
3.914
|
|
CCND1
|
cyclin D1
|
|
chr17_+_27369917
|
3.813
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr4_+_77610610
|
3.766
|
|
SHROOM3
|
shroom family member 3
|
|
chr5_-_39364562
|
3.740
|
NM_001737
|
C9
|
complement component 9
|
|
chr11_+_69455837
|
3.728
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr16_+_72088507
|
3.701
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr3_-_42917191
|
3.692
|
|
CYP8B1
|
cytochrome P450, family 8, subfamily B, polypeptide 1
|
|
chr8_-_26724728
|
3.607
|
|
|
|
|
chr12_+_20963637
|
3.584
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr13_-_86373328
|
3.543
|
NM_032229
|
SLITRK6
|
SLIT and NTRK-like family, member 6
|
|
chr2_-_216289936
|
3.480
|
|
FN1
|
fibronectin 1
|
|
chr10_+_7745354
|
3.432
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chr17_-_67138013
|
3.354
|
NM_080284
|
ABCA6
|
ATP-binding cassette, sub-family A (ABC1), member 6
|
|
chr2_+_169659099
|
3.338
|
NM_001039724
NM_001171632
NM_052946
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr10_+_96522457
|
3.332
|
NM_000769
|
CYP2C19
|
cytochrome P450, family 2, subfamily C, polypeptide 19
|
|
chr17_-_64225496
|
3.293
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr19_+_7828034
|
3.282
|
NM_001144904
NM_001144905
NM_001144906
NM_001144907
NM_001144908
NM_001144909
NM_001144910
NM_001144911
NM_014257
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr17_+_3379295
|
3.262
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr4_-_187644983
|
3.242
|
NM_005245
|
FAT1
|
FAT tumor suppressor homolog 1 (Drosophila)
|
|
chr9_-_104145800
|
3.214
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr3_+_153838791
|
3.189
|
NM_001251962
|
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26
|
|
chr7_+_136553398
|
3.119
|
NM_001006627
NM_001006630
|
CHRM2
|
cholinergic receptor, muscarinic 2
|
|
chr11_+_69455978
|
3.023
|
|
CCND1
|
cyclin D1
|
|
chr2_-_175712276
|
2.833
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr16_-_21314371
|
2.829
|
NM_001014444
NM_001888
|
CRYM
|
crystallin, mu
|
|
chr8_-_26724748
|
2.735
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr17_+_12623605
|
2.670
|
NM_001146313
|
MYOCD
|
myocardin
|
|
chr1_-_120311466
|
2.609
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr2_+_74120132
|
2.434
|
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr8_-_86253863
|
2.385
|
NM_001164830
|
CA1
|
carbonic anhydrase I
|
|
chr4_+_187187342
|
2.370
|
|
F11
|
coagulation factor XI
|
|
chr5_-_176836488
|
2.364
|
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr6_-_43276529
|
2.356
|
NM_206922
|
CRIP3
|
cysteine-rich protein 3
|
|
chr4_+_77610719
|
2.343
|
|
SHROOM3
|
shroom family member 3
|
|
chr2_+_74120092
|
2.333
|
NM_001199893
NM_001615
|
ACTG2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr1_+_57320442
|
2.275
|
NM_000562
|
C8A
|
complement component 8, alpha polypeptide
|
|
chr19_-_48389514
|
2.251
|
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1
|
|
chr7_-_16505292
|
2.250
|
NM_015464
|
SOSTDC1
|
sclerostin domain containing 1
|
|
chr3_-_9920633
|
2.229
|
NM_022094
|
CIDEC
|
cell death-inducing DFFA-like effector c
|
|
chr10_-_52645430
|
2.194
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr1_+_50574589
|
2.168
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr6_-_138820580
|
2.151
|
|
|
|
|
chr9_-_13250313
|
2.142
|
NM_003829
|
MPDZ
|
multiple PDZ domain protein
|
|
chr3_-_165555167
|
2.125
|
|
BCHE
|
butyrylcholinesterase
|
|
chr15_+_65337707
|
2.113
|
NM_178859
|
OSTBETA
|
organic solute transporter beta
|
|
chr10_-_96829074
|
2.109
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr4_+_77356252
|
2.098
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr7_-_16844606
|
2.096
|
NM_006408
|
AGR2
|
anterior gradient 2 homolog (Xenopus laevis)
|
|
chr2_+_234600320
|
2.075
|
NM_205862
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6
|
|
chr2_+_233734993
|
2.070
|
NM_206895
|
C2orf82
|
chromosome 2 open reading frame 82
|
|
chr13_-_28543290
|
2.006
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr12_-_14996381
|
1.993
|
NM_021071
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group)
|
|
chr4_+_184826427
|
1.979
|
NM_020225
|
STOX2
|
storkhead box 2
|
|
chr8_-_27115897
|
1.956
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr7_-_107443631
|
1.905
|
NM_000111
|
SLC26A3
|
solute carrier family 26, member 3
|
|
chr5_+_118965253
|
1.900
|
NM_001163991
NM_182761
|
FAM170A
|
family with sequence similarity 170, member A
|
|
chr2_-_231989755
|
1.893
|
NM_000867
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B
|
|
chr10_+_5005685
|
1.835
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr7_+_89843133
|
1.830
|
NM_001040666
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr3_-_42305438
|
1.828
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr4_-_110723334
|
1.826
|
NM_000204
|
CFI
|
complement factor I
|
|
chr1_+_163038934
|
1.825
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr9_-_140196702
|
1.818
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr5_+_173472692
|
1.798
|
NM_015980
|
HMP19
|
HMP19 protein
|
|
chr6_-_138820578
|
1.791
|
NM_001144060
|
NHSL1
|
NHS-like 1
|
|
chr17_+_32582295
|
1.778
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr5_-_135290521
|
1.775
|
NM_002302
|
LECT2
|
leukocyte cell-derived chemotaxin 2
|
|
chr12_-_21487831
|
1.764
|
NM_021094
|
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2
|
|
chr2_+_211458078
|
1.752
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr1_+_50574601
|
1.738
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_177133950
|
1.738
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr11_+_112047088
|
1.725
|
NM_001037290
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr8_+_76452202
|
1.718
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr14_+_39703119
|
1.716
|
NM_054024
|
MIA2
|
melanoma inhibitory activity 2
|
|
chr5_-_176836535
|
1.698
|
NM_000505
|
F12
|
coagulation factor XII (Hageman factor)
|
|
chr10_+_5005601
|
1.689
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr2_-_21225640
|
1.659
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr11_+_112046207
|
1.658
|
NM_031938
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr3_-_148939784
|
1.649
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr22_+_18043147
|
1.614
|
NM_031481
|
SLC25A18
|
solute carrier family 25 (mitochondrial carrier), member 18
|
|
chr1_+_196946666
|
1.609
|
NM_030787
|
CFHR5
|
complement factor H-related 5
|
|
chr1_+_101185195
|
1.600
|
NM_001078
NM_001199834
NM_080682
|
VCAM1
|
vascular cell adhesion molecule 1
|
|
chr11_-_118047336
|
1.599
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chr18_+_29171729
|
1.593
|
NM_000371
|
TTR
|
transthyretin
|
|
chr4_-_70361578
|
1.592
|
NM_021139
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4
|
|
chr4_+_74281884
|
1.585
|
|
ALB
|
albumin
|
|
chr13_-_28543143
|
1.559
|
|
CDX2
|
caudal type homeobox 2
|
|
chr14_-_106641585
|
1.541
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_5005453
|
1.532
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr1_+_87012758
|
1.514
|
NM_012128
|
CLCA4
|
chloride channel accessory 4
|
|
chr3_+_63428752
|
1.512
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr4_+_187187117
|
1.487
|
NM_000128
|
F11
|
coagulation factor XI
|
|
chr4_-_41750936
|
1.477
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr17_+_3377346
|
1.477
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr2_-_50574891
|
1.467
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr7_+_29519485
|
1.464
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr20_-_9819406
|
1.430
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr13_+_97794050
|
1.422
|
|
MBNL2
|
muscleblind-like 2 (Drosophila)
|
|
chr10_-_5045967
|
1.418
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr7_+_1126442
|
1.402
|
NM_001039966
NM_001505
|
GPER
|
G protein-coupled estrogen receptor 1
|
|
chr3_-_165555252
|
1.395
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr8_-_27115935
|
1.391
|
|
STMN4
|
stathmin-like 4
|
|
chr1_-_112106562
|
1.389
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chrX_+_138612894
|
1.380
|
NM_000133
|
F9
|
coagulation factor IX
|
|
chr19_-_5838740
|
1.377
|
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr10_-_69597768
|
1.366
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr3_-_93747453
|
1.361
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr5_+_176516550
|
1.350
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chr18_-_61329113
|
1.340
|
NM_006919
|
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3
|
|
chrX_-_15402469
|
1.327
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr12_+_21284127
|
1.308
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr1_+_81771844
|
1.302
|
|
LPHN2
|
latrophilin 2
|
|
chr9_+_97488993
|
1.299
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr3_-_52864687
|
1.293
|
NM_001166449
NM_002218
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4
|
|
chr13_-_28543423
|
1.273
|
|
CDX2
|
caudal type homeobox 2
|
|
chr18_-_3845295
|
1.273
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr16_+_76311175
|
1.252
|
NM_033401
|
CNTNAP4
|
contactin associated protein-like 4
|
|
chr1_+_196912910
|
1.233
|
NM_005666
|
CFHR2
|
complement factor H-related 2
|
|
chr3_-_170744767
|
1.231
|
NM_000340
|
SLC2A2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chrX_+_144908927
|
1.229
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chr13_-_110802708
|
1.201
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr4_-_155511838
|
1.177
|
NM_000508
NM_021871
|
FGA
|
fibrinogen alpha chain
|
|
chr3_+_2933898
|
1.172
|
NM_001206956
NM_175613
|
CNTN4
|
contactin 4
|
|
chr9_-_74675498
|
1.170
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr4_-_176733413
|
1.158
|
|
GPM6A
|
glycoprotein M6A
|
|
chr6_+_131894343
|
1.157
|
NM_000045
NM_001244438
|
ARG1
|
arginase, liver
|
|
chr2_+_182850550
|
1.156
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr5_+_173472737
|
1.153
|
|
HMP19
|
HMP19 protein
|
|
chr3_+_100238063
|
1.153
|
|
TMEM45A
|
transmembrane protein 45A
|
|
chr7_-_8302141
|
1.152
|
NM_004968
|
ICA1
|
islet cell autoantigen 1, 69kDa
|
|
chr14_-_107170400
|
1.141
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr6_+_153071931
|
1.139
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr2_-_224467142
|
1.137
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr12_-_102874373
|
1.125
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_-_42908178
|
1.118
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr11_+_63057371
|
1.109
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr3_+_89156673
|
1.108
|
NM_005233
NM_182644
|
EPHA3
|
EPH receptor A3
|
|
chr12_-_102874340
|
1.106
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_+_41831484
|
1.105
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr5_+_140771482
|
1.103
|
NM_014004
NM_032088
|
PCDHGA8
|
protocadherin gamma subfamily A, 8
|
|
chr11_-_18258315
|
1.088
|
|
SAA4
|
serum amyloid A4, constitutive
|
|
chr14_-_106733392
|
1.074
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr2_-_216242916
|
1.067
|
|
FN1
|
fibronectin 1
|
|
chr8_-_120651019
|
1.063
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr19_-_51961648
|
1.051
|
NM_014442
|
SIGLEC8
|
sialic acid binding Ig-like lectin 8
|
|
chr4_-_41750722
|
1.051
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr2_+_113885145
|
1.050
|
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr3_+_108021331
|
1.046
|
NM_007072
|
HHLA2
|
HERV-H LTR-associating 2
|
|
chr2_+_166326156
|
1.044
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr4_-_176733378
|
1.041
|
|
GPM6A
|
glycoprotein M6A
|
|
chrX_-_139047676
|
1.038
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr1_+_16083132
|
1.036
|
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr19_-_5839702
|
1.034
|
NM_000150
NM_001040701
|
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase)
|
|
chr14_-_106452693
|
1.021
|
|
IGHG1
|
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr1_+_162602198
|
1.016
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr2_+_234826042
|
1.013
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr3_+_2280512
|
1.013
|
NM_001206955
|
CNTN4
|
contactin 4
|
|
chr12_-_102874158
|
1.013
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr12_-_27599557
|
0.992
|
|
ARNTL2
|
aryl hydrocarbon receptor nuclear translocator-like 2
|
|
chr7_-_122339186
|
0.985
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr4_-_122085475
|
0.980
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr5_+_125758818
|
0.971
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_+_135596185
|
0.967
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr10_+_7745341
|
0.963
|
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2
|
|
chrX_+_142113703
|
0.958
|
NM_001009613
|
SPANXN4
|
SPANX family, member N4
|
|
chr18_-_28622624
|
0.957
|
NM_001941
NM_024423
|
DSC3
|
desmocollin 3
|
|
chr19_+_56270506
|
0.949
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr5_+_161277702
|
0.947
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_186578122
|
0.946
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr19_+_42259397
|
0.935
|
NM_002483
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen)
|
|
chr18_-_58040000
|
0.924
|
NM_005912
|
MC4R
|
melanocortin 4 receptor
|
|
chr15_+_22736245
|
0.915
|
NM_001001413
|
GOLGA6L1
|
golgin A6 family-like 1
|
|
chr6_-_49754899
|
0.901
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr18_+_46065367
|
0.886
|
NM_001142397
NM_014772
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr2_+_90077732
|
0.881
|
|
|
|
|
chr3_-_149095413
|
0.870
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr10_+_114133775
|
0.867
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr3_+_159557649
|
0.866
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_-_8815370
|
0.862
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr9_+_97521930
|
0.862
|
NM_001193329
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr11_+_60524339
|
0.861
|
NM_001098835
NM_152717
|
MS4A15
|
membrane-spanning 4-domains, subfamily A, member 15
|
|
chr4_-_6422844
|
0.859
|
NM_001206996
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr12_+_18891040
|
0.854
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr9_+_131684561
|
0.849
|
NM_001100877
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr9_+_74729510
|
0.824
|
NM_001242507
|
GDA
|
guanine deaminase
|