|
chr11_-_26593667
|
7.359
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr8_-_49833823
|
5.241
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr9_+_34458810
|
5.047
|
NM_012144
|
DNAI1
|
dynein, axonemal, intermediate chain 1
|
|
chr16_-_31439650
|
4.782
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr9_-_95166827
|
4.621
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chrX_+_65382432
|
3.977
|
NM_138737
|
HEPH
|
hephaestin
|
|
chr8_+_17396285
|
3.868
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr7_+_128349111
|
3.836
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr10_-_30918625
|
3.768
|
NM_183058
|
LYZL2
|
lysozyme-like 2
|
|
chrX_+_150884506
|
3.653
|
NM_033085
|
FATE1
|
fetal and adult testis expressed 1
|
|
chr1_-_116311398
|
3.498
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr5_-_137475076
|
3.493
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr5_+_140529619
|
3.461
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr1_-_116311161
|
3.400
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311330
|
3.378
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_67594219
|
3.306
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr13_+_43355685
|
3.199
|
NM_182508
|
C13orf30
|
chromosome 13 open reading frame 30
|
|
chr6_-_123957941
|
3.181
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr10_+_29577989
|
3.100
|
NM_032517
|
LYZL1
|
lysozyme-like 1
|
|
chr18_+_47088400
|
3.069
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr19_-_49658645
|
2.979
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr8_+_70378858
|
2.833
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr6_+_47749774
|
2.821
|
NM_181744
|
OPN5
|
opsin 5
|
|
chr12_-_7848324
|
2.748
|
NM_020634
|
GDF3
|
growth differentiation factor 3
|
|
chr1_+_180199432
|
2.690
|
NM_033343
|
LHX4
|
LIM homeobox 4
|
|
chr13_-_20110884
|
2.612
|
NM_001141968
NM_130785
NM_199254
|
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2
|
|
chr1_-_76397975
|
2.601
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr5_-_156593178
|
2.540
|
NM_130899
|
FAM71B
|
family with sequence similarity 71, member B
|
|
chr15_+_22382381
|
2.520
|
NM_001005241
|
OR4N4
|
olfactory receptor, family 4, subfamily N, member 4
|
|
chr9_+_35041101
|
2.451
|
NM_001040411
NM_001040412
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr10_-_128209996
|
2.430
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr6_+_105404922
|
2.410
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr5_+_150040402
|
2.398
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr2_-_27362277
|
2.292
|
NM_178553
|
C2orf53
|
chromosome 2 open reading frame 53
|
|
chr14_-_60952734
|
2.251
|
NM_174978
|
C14orf39
|
chromosome 14 open reading frame 39
|
|
chr17_-_10421858
|
2.221
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr22_-_22863504
|
2.182
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr11_-_83393436
|
2.158
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr20_+_138121
|
2.127
|
NM_139074
|
DEFB127
|
defensin, beta 127
|
|
chr5_+_140602914
|
2.085
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr6_-_49834188
|
2.071
|
NM_001131
NM_170609
|
CRISP1
|
cysteine-rich secretory protein 1
|
|
chr15_+_54305100
|
2.063
|
NM_001080534
|
UNC13C
|
unc-13 homolog C (C. elegans)
|
|
chr1_+_84768333
|
2.033
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr7_+_94297448
|
2.029
|
|
PEG10
|
paternally expressed 10
|
|
chr8_+_70379077
|
1.995
|
|
SULF1
|
sulfatase 1
|
|
chr11_+_57310113
|
1.988
|
NM_001105565
|
SMTNL1
|
smoothelin-like 1
|
|
chr3_-_169487602
|
1.949
|
NM_032487
|
ARPM1
|
actin related protein M1
|
|
chr17_-_67240955
|
1.889
|
NM_080282
|
ABCA10
|
ATP-binding cassette, sub-family A (ABC1), member 10
|
|
chr21_+_19289656
|
1.870
|
NM_001204175
NM_001204176
NM_001204177
NM_001204178
|
CHODL
|
chondrolectin
|
|
chr7_-_27183225
|
1.849
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr5_+_92918924
|
1.790
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr2_+_233390869
|
1.778
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr7_-_100026284
|
1.750
|
NM_017984
|
ZCWPW1
|
zinc finger, CW type with PWWP domain 1
|
|
chr4_-_44653657
|
1.726
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr4_+_78432905
|
1.722
|
NM_006419
|
CXCL13
|
chemokine (C-X-C motif) ligand 13
|
|
chr2_-_180427314
|
1.717
|
NM_001113398
|
ZNF385B
|
zinc finger protein 385B
|
|
chr8_+_36641841
|
1.708
|
NM_001031836
|
KCNU1
|
potassium channel, subfamily U, member 1
|
|
chr19_-_53794874
|
1.677
|
NM_033341
|
BIRC8
|
baculoviral IAP repeat containing 8
|
|
chr15_+_49715374
|
1.626
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr9_-_16727887
|
1.611
|
|
BNC2
|
basonuclin 2
|
|
chr4_+_41614911
|
1.604
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_-_36236421
|
1.512
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr4_-_152149042
|
1.506
|
NM_001243349
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_-_91572328
|
1.503
|
NM_133504
NM_133505
NM_133506
NM_133507
|
DCN
|
decorin
|
|
chr10_+_69869249
|
1.475
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr4_-_46125929
|
1.470
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr9_-_95298251
|
1.463
|
NM_001197295
NM_001197296
NM_001393
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr6_-_26108320
|
1.413
|
NM_005323
|
HIST1H1T
|
histone cluster 1, H1t
|
|
chr3_+_46919235
|
1.412
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr1_+_109255555
|
1.412
|
NM_001144937
|
FNDC7
|
fibronectin type III domain containing 7
|
|
chr2_-_72371188
|
1.386
|
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr8_-_125183703
|
1.381
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chrX_+_65382627
|
1.371
|
|
HEPH
|
hephaestin
|
|
chr11_+_5509914
|
1.367
|
NM_001005163
|
OR52D1
|
olfactory receptor, family 52, subfamily D, member 1
|
|
chr3_+_127641901
|
1.357
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr19_-_22379752
|
1.354
|
NM_001001411
|
ZNF676
|
zinc finger protein 676
|
|
chr17_-_10325266
|
1.325
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr20_+_123230
|
1.313
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr14_-_94442894
|
1.286
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr19_-_56249739
|
1.281
|
NM_176820
|
NLRP9
|
NLR family, pyrin domain containing 9
|
|
chrX_-_33229428
|
1.261
|
NM_004006
|
DMD
|
dystrophin
|
|
chr7_+_94292737
|
1.243
|
|
PEG10
|
paternally expressed 10
|
|
chr7_+_134464372
|
1.238
|
|
CALD1
|
caldesmon 1
|
|
chr5_-_156390206
|
1.208
|
NM_001146726
NM_138379
|
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4
|
|
chr3_-_185826748
|
1.192
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr14_+_51955854
|
1.181
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr4_+_175839508
|
1.165
|
NM_001130703
NM_001130704
NM_001130705
NM_014269
|
ADAM29
|
ADAM metallopeptidase domain 29
|
|
chr7_+_119913688
|
1.164
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr15_+_74422742
|
1.143
|
NM_020851
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr6_+_53883713
|
1.138
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chrX_+_135278912
|
1.124
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr15_-_31393906
|
1.113
|
NM_001252024
NM_001252030
NM_002420
|
TRPM1
|
transient receptor potential cation channel, subfamily M, member 1
|
|
chr22_-_40289793
|
1.103
|
NM_152512
|
ENTHD1
|
ENTH domain containing 1
|
|
chr3_+_128199744
|
1.102
|
|
|
|
|
chr3_-_151158558
|
1.092
|
NM_001178145
NM_001178146
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr5_+_125758818
|
1.091
|
NM_001146320
NM_001146321
NM_023927
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_-_183387063
|
1.088
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr5_+_42756919
|
1.078
|
NM_001134848
|
CCDC152
|
coiled-coil domain containing 152
|
|
chr18_-_51880942
|
1.070
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr2_+_166326156
|
1.068
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr9_+_108424737
|
1.060
|
NM_005421
|
TAL2
|
T-cell acute lymphocytic leukemia 2
|
|
chr11_-_16497917
|
1.047
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr12_-_15865847
|
1.040
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr6_+_152011630
|
1.036
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr10_+_5005453
|
1.030
|
NM_001353
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr10_+_68685791
|
1.023
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr12_-_59314069
|
1.018
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr1_+_215178884
|
1.007
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr4_+_119948322
|
1.005
|
|
SYNPO2
|
synaptopodin 2
|
|
chr3_-_111314181
|
1.004
|
NM_024508
|
ZBED2
|
zinc finger, BED-type containing 2
|
|
chr12_-_28124915
|
1.004
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr20_-_50313990
|
1.004
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr11_-_57417247
|
0.989
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr4_-_65275000
|
0.985
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr6_-_49754899
|
0.981
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr1_+_160336856
|
0.978
|
NM_005598
|
NHLH1
|
nescient helix loop helix 1
|
|
chr4_-_87028805
|
0.963
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr3_-_151987332
|
0.962
|
|
LOC401093
|
uncharacterized LOC401093
|
|
chr16_+_81272293
|
0.951
|
NM_017429
|
BCMO1
|
beta-carotene 15,15'-monooxygenase 1
|
|
chr22_-_36236264
|
0.948
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr12_+_100041527
|
0.946
|
NM_153364
|
FAM71C
|
family with sequence similarity 71, member C
|
|
chr10_+_5005685
|
0.939
|
|
AKR1C1
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
|
|
chr7_-_27159213
|
0.933
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr17_-_66951381
|
0.928
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr6_+_46714653
|
0.918
|
NM_001162435
|
LOC100287718
|
uncharacterized LOC100287718
|
|
chr4_+_119948554
|
0.911
|
|
SYNPO2
|
synaptopodin 2
|
|
chr18_+_61144143
|
0.909
|
NM_002639
|
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5
|
|
chr17_+_3379295
|
0.909
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr8_-_93075190
|
0.892
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_-_7218370
|
0.891
|
NM_004489
|
GPS2
|
G protein pathway suppressor 2
|
|
chr1_+_196621007
|
0.891
|
NM_000186
NM_001014975
|
CFH
|
complement factor H
|
|
chr14_-_21058880
|
0.882
|
NM_001024822
|
RNASE12
|
ribonuclease, RNase A family, 12 (non-active)
|
|
chr16_-_32688052
|
0.879
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr1_+_26503973
|
0.862
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chrX_+_86772714
|
0.858
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr13_-_103411421
|
0.848
|
NM_001146197
|
CCDC168
|
coiled-coil domain containing 168
|
|
chr8_+_87111132
|
0.838
|
NM_152565
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2
|
|
chr17_-_10372875
|
0.837
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr10_+_5005601
|
0.823
|
|
AKR1C1
AKR1C3
|
aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase)
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr17_-_54893249
|
0.807
|
NM_001085430
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr14_-_101351183
|
0.802
|
NM_001134888
|
RTL1
|
retrotransposon-like 1
|
|
chrX_-_107682600
|
0.800
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr19_+_58180302
|
0.771
|
NM_152677
|
ZSCAN4
|
zinc finger and SCAN domain containing 4
|
|
chr4_+_62362838
|
0.761
|
NM_015236
|
LPHN3
|
latrophilin 3
|
|
chr8_-_107782417
|
0.760
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr14_+_51026742
|
0.757
|
NM_015915
NM_181598
|
ATL1
|
atlastin GTPase 1
|
|
chr8_-_12973752
|
0.746
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr8_-_93107881
|
0.743
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_51611476
|
0.742
|
|
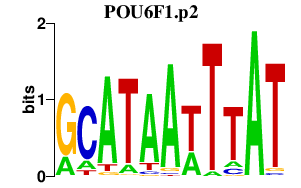
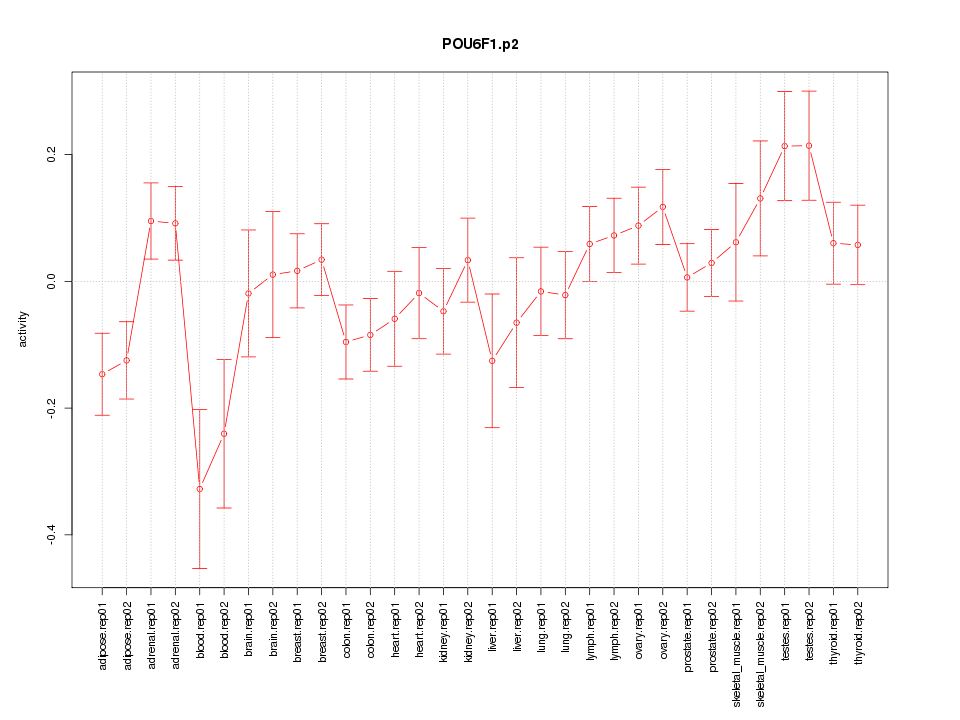
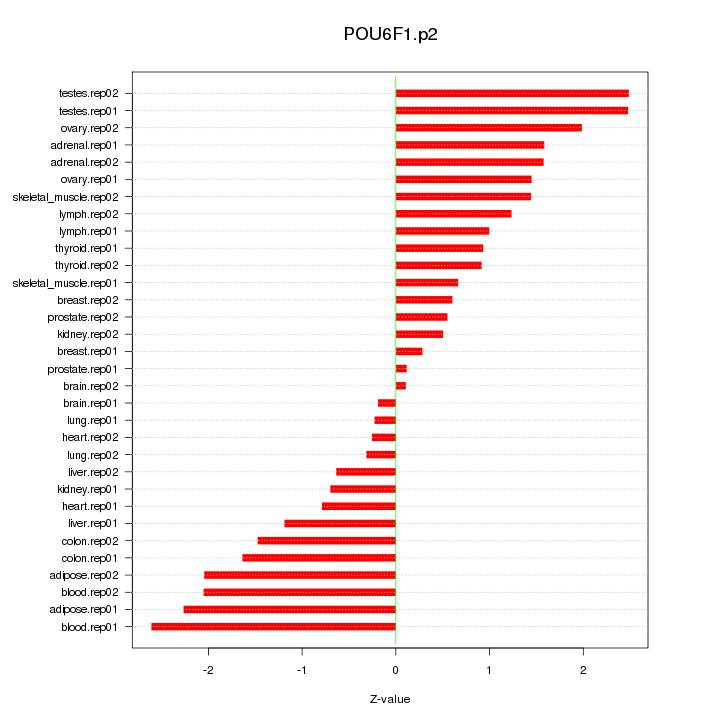
POU6F1
|
POU class 6 homeobox 1
|
|
chr17_+_17608163
|
0.741
|
|
|
|
|
chr1_+_15272414
|
0.740
|
NM_001017999
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chrX_-_139866582
|
0.736
|
NM_004065
|
CDR1
|
cerebellar degeneration-related protein 1, 34kDa
|
|
chr1_+_46379258
|
0.735
|
|
MAST2
|
microtubule associated serine/threonine kinase 2
|
|
chr4_-_152147520
|
0.735
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr10_-_72545110
|
0.727
|
NM_152710
|
C10orf27
|
chromosome 10 open reading frame 27
|
|
chr6_-_112575708
|
0.720
|
|
LAMA4
|
laminin, alpha 4
|
|
chr17_+_41857802
|
0.719
|
NM_001136483
|
C17orf105
|
chromosome 17 open reading frame 105
|
|
chr14_-_57960432
|
0.716
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr7_+_138818489
|
0.709
|
NM_001144920
NM_001144923
NM_024926
|
TTC26
|
tetratricopeptide repeat domain 26
|
|
chr10_-_5045967
|
0.705
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr4_+_41614725
|
0.704
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_+_9479943
|
0.704
|
NM_001080556
NM_145054
|
WDR16
|
WD repeat domain 16
|
|
chrX_+_47986602
|
0.701
|
NM_205856
|
SPACA5
|
sperm acrosome associated 5
|
|
chr10_-_43187174
|
0.694
|
|
|
|
|
chr11_+_22696318
|
0.693
|
NM_005256
|
GAS2
|
growth arrest-specific 2
|
|
chrX_+_90689596
|
0.692
|
NM_080832
|
PABPC5
|
poly(A) binding protein, cytoplasmic 5
|
|
chr11_-_8190535
|
0.691
|
NM_001135109
NM_001206671
NM_001206672
NM_024557
|
RIC3
|
resistance to inhibitors of cholinesterase 3 homolog (C. elegans)
|
|
chrX_+_91090459
|
0.687
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr7_+_142829169
|
0.680
|
NM_002652
|
PIP
|
prolactin-induced protein
|
|
chr11_-_102401430
|
0.667
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr13_+_24144402
|
0.666
|
NM_001204459
NM_148957
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19
|
|
chr4_+_166794409
|
0.666
|
NM_001204760
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chrX_-_117119282
|
0.663
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr1_-_163113938
|
0.662
|
|
RGS5
|
regulator of G-protein signaling 5
|
|
chr10_+_15001437
|
0.661
|
NM_001080836
|
MEIG1
|
meiosis expressed gene 1 homolog (mouse)
|
|
chr15_+_74422920
|
0.660
|
|
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2
|
|
chr4_-_122085475
|
0.660
|
NM_024873
|
TNIP3
|
TNFAIP3 interacting protein 3
|
|
chr12_-_59314297
|
0.657
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr14_+_70918873
|
0.655
|
NM_003813
|
ADAM21
|
ADAM metallopeptidase domain 21
|
|
chr10_-_13544944
|
0.646
|
NM_001100912
NM_152751
|
BEND7
|
BEN domain containing 7
|
|
chr1_+_84874033
|
0.637
|
NM_058248
|
DNASE2B
|
deoxyribonuclease II beta
|
|
chr5_+_125759076
|
0.635
|
|
GRAMD3
|
GRAM domain containing 3
|
|
chr2_-_180610768
|
0.633
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chrX_+_36254050
|
0.628
|
NM_001098843
|
CXorf30
|
chromosome X open reading frame 30
|
|
chr19_+_110624
|
0.618
|
NM_001005240
|
OR4F4
OR4F17
|
olfactory receptor, family 4, subfamily F, member 4
olfactory receptor, family 4, subfamily F, member 17
|
|
chr5_+_140220811
|
0.617
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr3_-_57234279
|
0.616
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr4_+_41614933
|
0.606
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chrX_-_32173585
|
0.604
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr1_-_117746457
|
0.593
|
NM_001253849
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chrX_-_15402469
|
0.584
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr1_+_18081807
|
0.582
|
NM_030812
|
ACTL8
|
actin-like 8
|
|
chr18_+_616671
|
0.582
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr12_-_11150473
|
0.580
|
NM_176889
|
TAS2R20
|
taste receptor, type 2, member 20
|
|
chr16_+_33204979
|
0.575
|
NM_016212
|
TP53TG3
|
TP53 target 3
|
|
chr2_-_56151273
|
0.573
|
NM_001039348
NM_001039349
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1
|
|
chr10_-_17171789
|
0.571
|
NM_001081
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr11_-_27681195
|
0.569
|
NM_001143816
NM_170735
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_-_104238785
|
0.568
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr12_+_54426831
|
0.555
|
NM_018953
|
HOXC5
|
homeobox C5
|