|
chr9_-_34710062
|
13.854
|
NM_002989
|
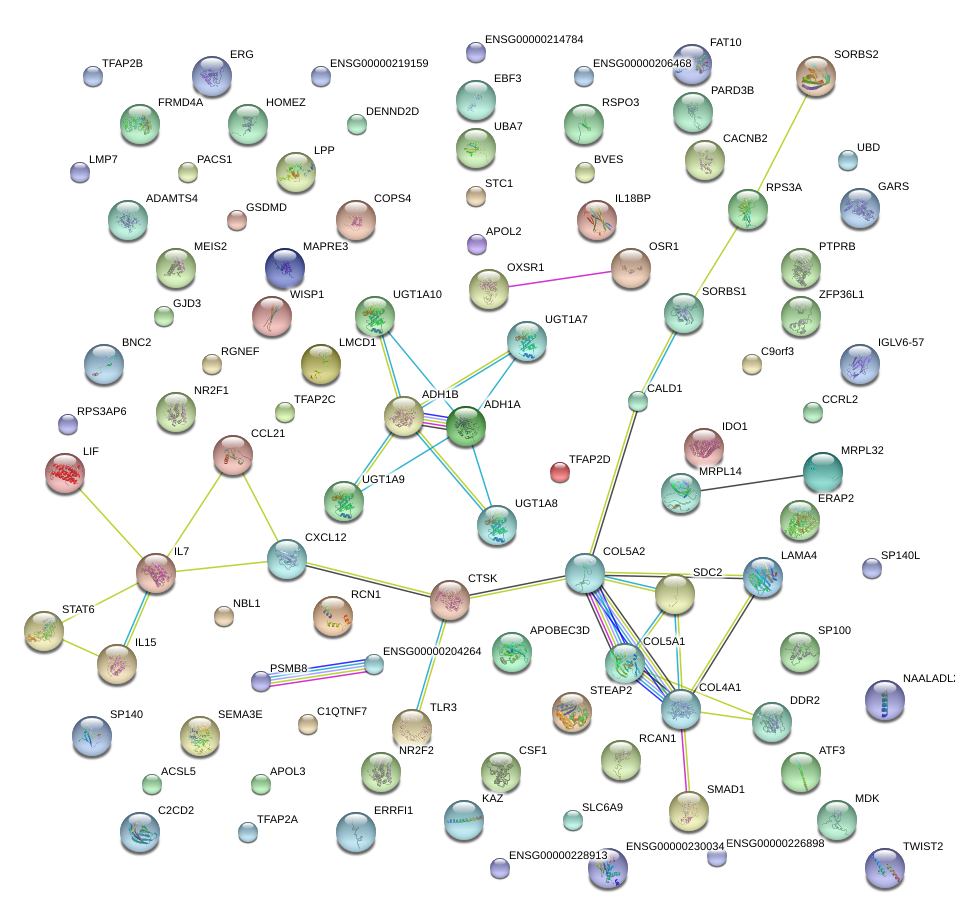
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr10_-_44880452
|
11.685
|
NM_000609
NM_001033886
NM_001178134
NM_199168
|
CXCL12
|
chemokine (C-X-C motif) ligand 12
|
|
chr6_-_112575766
|
10.893
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_112575708
|
9.410
|
|
LAMA4
|
laminin, alpha 4
|
|
chr4_-_100242487
|
9.065
|
NM_000668
|
ADH1A
ADH1B
|
alcohol dehydrogenase 1A (class I), alpha polypeptide
alcohol dehydrogenase 1B (class I), beta polypeptide
|
|
chr2_+_239756672
|
8.604
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr6_-_112575345
|
7.451
|
|
LAMA4
|
laminin, alpha 4
|
|
chr10_-_131762017
|
6.843
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr6_-_112575880
|
6.754
|
|
LAMA4
|
laminin, alpha 4
|
|
chr6_-_29527477
|
6.467
|
|
UBD
|
ubiquitin D
|
|
chr1_+_19971865
|
6.280
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr2_-_190044330
|
6.066
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_+_134203281
|
6.036
|
NM_001204869
NM_001204870
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr15_+_96876568
|
5.964
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr22_-_30642683
|
5.481
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_+_19972259
|
5.201
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_+_212781969
|
5.059
|
NM_001040619
NM_001206484
NM_001206488
NM_001674
|
ATF3
|
activating transcription factor 3
|
|
chr6_-_29527701
|
4.972
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr9_-_16727887
|
4.823
|
|
BNC2
|
basonuclin 2
|
|
chr1_+_212782103
|
4.790
|
|
ATF3
|
activating transcription factor 3
|
|
chr6_+_127440047
|
4.600
|
NM_032784
|
RSPO3
|
R-spondin 3
|
|
chr3_+_8543508
|
4.373
|
NM_014583
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr12_-_71003622
|
4.363
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr6_+_127439803
|
4.318
|
|
RSPO3
|
R-spondin 3
|
|
chr4_+_186990308
|
4.297
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr15_-_37393364
|
4.263
|
NM_001220482
NM_002399
|
MEIS2
|
Meis homeobox 2
|
|
chr1_+_110453456
|
4.235
|
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr3_+_8543539
|
4.156
|
|
LMCD1
|
LIM and cysteine-rich domains 1
|
|
chr1_-_44497132
|
4.118
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr1_+_110453232
|
4.105
|
NM_000757
NM_172210
NM_172212
|
CSF1
|
colony stimulating factor 1 (macrophage)
|
|
chr22_+_22550159
|
4.033
|
|
IGLV6-57
|
immunoglobulin lambda variable 6-57
|
|
chr8_+_39771327
|
3.920
|
NM_002164
|
IDO1
|
indoleamine 2,3-dioxygenase 1
|
|
chr1_+_162602308
|
3.596
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr13_-_110802708
|
3.556
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_-_19558371
|
3.374
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr8_+_144640476
|
3.165
|
NM_024736
|
GSDMD
|
gasdermin D
|
|
chr9_+_137533643
|
2.973
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr14_-_69262959
|
2.826
|
NM_001244701
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr22_-_36556781
|
2.822
|
NM_145640
|
APOL3
|
apolipoprotein L, 3
|
|
chr1_-_44497028
|
2.811
|
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr1_+_162602198
|
2.782
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr4_-_186578122
|
2.678
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_+_97506020
|
2.647
|
|
SDC2
|
syndecan 2
|
|
chr12_-_57505176
|
2.629
|
NM_001178078
NM_001178080
NM_001178081
NM_003153
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr10_-_97321094
|
2.613
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr17_+_17398334
|
2.599
|
|
|
|
|
chr1_-_161168816
|
2.555
|
NM_005099
|
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif, 4
|
|
chr8_+_97505876
|
2.505
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr8_-_79717540
|
2.494
|
NM_000880
NM_001199886
NM_001199887
NM_001199888
|
IL7
|
interleukin 7
|
|
chr22_+_22550112
|
2.486
|
|
|
|
|
chr1_-_8075678
|
2.354
|
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr11_+_46402333
|
2.328
|
NM_001012334
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr2_+_205410716
|
2.268
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr2_+_205410465
|
2.123
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr3_-_49851217
|
2.103
|
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr9_+_97766414
|
2.066
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr10_+_114135965
|
2.027
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr14_-_69259654
|
2.003
|
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr2_+_231191877
|
2.001
|
NM_138402
|
SP140L
|
SP140 nuclear body protein-like
|
|
chr8_+_97505895
|
1.994
|
|
SDC2
|
syndecan 2
|
|
chr12_-_57505121
|
1.920
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr2_+_234526290
|
1.874
|
NM_019076
|
UGT1A8
|
UDP glucuronosyltransferase 1 family, polypeptide A8
|
|
chr8_+_97506071
|
1.858
|
|
SDC2
|
syndecan 2
|
|
chr10_+_114135937
|
1.828
|
NM_016234
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr22_+_39417117
|
1.824
|
NM_152426
|
APOBEC3D
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D
|
|
chr3_-_49851356
|
1.758
|
NM_003335
|
UBA7
|
ubiquitin-like modifier activating enzyme 7
|
|
chr5_+_96212167
|
1.687
|
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2
|
|
chr17_-_38520941
|
1.682
|
NM_152219
|
GJD3
|
gap junction protein, delta 3, 31.9kDa
|
|
chr11_+_32112607
|
1.676
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr21_-_43346780
|
1.667
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr4_+_15376174
|
1.660
|
NM_001135171
|
C1QTNF7
|
C1q and tumor necrosis factor related protein 7
|
|
chr6_-_10412606
|
1.637
|
NM_001032280
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr7_-_83270746
|
1.582
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr8_-_23712297
|
1.574
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr1_-_111746917
|
1.567
|
|
DENND2D
|
DENN/MADD domain containing 2D
|
|
chr7_+_134464161
|
1.506
|
NM_004342
NM_033138
NM_033157
|
CALD1
|
caldesmon 1
|
|
chr12_-_57504935
|
1.503
|
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced
|
|
chr21_-_40033617
|
1.501
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr6_-_105584542
|
1.473
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr4_-_186577858
|
1.449
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_-_13749622
|
1.408
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_+_231280933
|
1.402
|
|
SP100
|
SP100 nuclear antigen
|
|
chr1_-_150780705
|
1.373
|
NM_000396
|
CTSK
|
cathepsin K
|
|
chr4_+_152020732
|
1.360
|
NM_001006
|
RPS3A
|
ribosomal protein S3A
|
|
chr2_+_231280906
|
1.360
|
|
SP100
|
SP100 nuclear antigen
|
|
chr2_+_231280870
|
1.358
|
NM_001080391
NM_001206701
NM_001206702
NM_001206703
NM_003113
|
SP100
|
SP100 nuclear antigen
|
|
chr10_+_114136014
|
1.352
|
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5
|
|
chr11_+_65190272
|
1.333
|
|
|
|
|
chr7_+_42971938
|
1.330
|
NM_031903
|
MRPL32
|
mitochondrial ribosomal protein L32
|
|
chr7_+_89843133
|
1.321
|
NM_001040666
|
STEAP2
|
STEAP family member 2, metalloreductase
|
|
chr11_+_71709951
|
1.313
|
NM_001039659
NM_001039660
NM_173044
NM_005699
NM_173042
|
IL18BP
|
interleukin 18 binding protein
|
|
chr2_+_205410556
|
1.280
|
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr2_+_234545099
|
1.279
|
NM_019075
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10
|
|
chr11_+_71710971
|
1.278
|
NM_001145055
|
IL18BP
|
interleukin 18 binding protein
|
|
chr11_+_65190267
|
1.277
|
|
NEAT1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr5_+_73109342
|
1.257
|
NM_001244364
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr3_+_46448678
|
1.242
|
NM_003965
NM_001130910
|
CCRL2
|
chemokine (C-C motif) receptor-like 2
|
|
chr6_-_32811773
|
1.232
|
NM_148919
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr10_+_18429605
|
1.226
|
NM_201593
NM_201596
NM_201597
NM_001167945
NM_201571
NM_201572
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr4_+_142557722
|
1.203
|
NM_000585
NM_172175
|
IL15
|
interleukin 15
|
|
chr14_-_69260630
|
1.203
|
NM_001244698
NM_004926
|
ZFP36L1
|
zinc finger protein 36, C3H type-like 1
|
|
chr3_+_174577069
|
1.186
|
NM_207015
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2
|
|
chr3_+_187871473
|
1.173
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr4_+_146403955
|
1.136
|
NM_001003688
|
SMAD1
|
SMAD family member 1
|
|
chr17_-_48271365
|
1.104
|
|
|
|
|
chr22_+_39436608
|
1.103
|
NM_001006666
NM_145298
|
APOBEC3F
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F
|
|
chr4_-_100140330
|
1.078
|
NM_000672
NM_001102470
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr10_+_102123420
|
1.053
|
|
|
|
|
chr19_+_49977465
|
1.039
|
NM_001204502
NM_001459
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr9_-_117568292
|
1.025
|
NM_005118
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr19_+_13105970
|
1.004
|
NM_002501
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor)
|
|
chr6_-_105584982
|
0.985
|
NM_001199563
|
BVES
|
blood vessel epicardial substance
|
|
chr18_-_53071225
|
0.985
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr6_+_36973409
|
0.966
|
NM_173558
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2
|
|
chr16_+_57023411
|
0.958
|
|
NLRC5
|
NLR family, CARD domain containing 5
|
|
chr12_+_102271349
|
0.944
|
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chrX_+_102883647
|
0.919
|
NM_001006640
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr11_+_71710662
|
0.910
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr9_+_109625347
|
0.882
|
NM_021224
|
ZNF462
|
zinc finger protein 462
|
|
chr13_+_108921976
|
0.871
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr12_+_102271082
|
0.863
|
NM_018370
|
DRAM1
|
DNA-damage regulated autophagy modulator 1
|
|
chr10_+_123754141
|
0.862
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr2_+_132480063
|
0.817
|
NM_013310
|
C2orf27A
|
chromosome 2 open reading frame 27A
|
|
chr16_-_85145995
|
0.817
|
NM_198491
|
FAM92B
|
family with sequence similarity 92, member B
|
|
chr10_+_63808968
|
0.797
|
NM_001244638
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr6_-_32806545
|
0.787
|
NM_000544
NM_018833
|
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr9_+_112542496
|
0.759
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr6_-_99797530
|
0.754
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr19_+_49977817
|
0.745
|
NM_001204503
|
FLT3LG
|
fms-related tyrosine kinase 3 ligand
|
|
chr17_+_41158741
|
0.737
|
NM_005533
|
IFI35
|
interferon-induced protein 35
|
|
chr1_-_57045235
|
0.716
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr17_+_6659155
|
0.697
|
NM_017523
NM_199139
|
XAF1
|
XIAP associated factor 1
|
|
chr21_+_34144410
|
0.690
|
|
C21orf49
|
chromosome 21 open reading frame 49
|
|
chr1_+_159796478
|
0.689
|
NM_020125
|
SLAMF8
|
SLAM family member 8
|
|
chr5_-_56778553
|
0.686
|
NM_001017992
|
ACTBL2
|
actin, beta-like 2
|
|
chr13_+_108922205
|
0.667
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr9_+_18474078
|
0.659
|
NM_001040272
NM_052866
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr15_-_82641705
|
0.639
|
NM_001164465
|
GOLGA6L10
|
golgin A6 family-like 10
|
|
chr9_+_18474138
|
0.633
|
|
ADAMTSL1
|
ADAMTS-like 1
|
|
chr7_+_134464372
|
0.621
|
|
CALD1
|
caldesmon 1
|
|
chr6_-_31704272
|
0.617
|
|
CLIC1
|
chloride intracellular channel 1
|
|
chr4_-_120243257
|
0.615
|
NM_000134
|
FABP2
|
fatty acid binding protein 2, intestinal
|
|
chr2_-_231084600
|
0.609
|
NM_004509
NM_004510
NM_080424
|
SP110
|
SP110 nuclear body protein
|
|
chr9_-_14868974
|
0.598
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr1_+_221052845
|
0.596
|
|
HLX
|
H2.0-like homeobox
|
|
chr8_-_42065037
|
0.589
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr22_+_39473009
|
0.588
|
NM_021822
|
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G
|
|
chrX_+_102883891
|
0.581
|
NM_001006639
NM_004780
|
TCEAL1
|
transcription elongation factor A (SII)-like 1
|
|
chr2_+_3627725
|
0.580
|
|
RPS7
|
ribosomal protein S7
|
|
chr16_-_73092533
|
0.565
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr6_+_32821924
|
0.562
|
NM_002800
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2)
|
|
chr6_-_33281847
|
0.555
|
|
TAPBP
|
TAP binding protein (tapasin)
|
|
chr8_-_93075190
|
0.552
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_+_69915374
|
0.551
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_+_83098709
|
0.546
|
NM_198181
|
GOLGA6L9
|
golgin A6 family-like 9
|
|
chr1_+_2985730
|
0.545
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr8_-_23540401
|
0.542
|
NM_006167
|
NKX3-1
|
NK3 homeobox 1
|
|
chr14_+_24701624
|
0.535
|
NM_001002000
NM_001002001
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr5_+_32710742
|
0.531
|
NM_001204376
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C)
|
|
chr2_+_121554866
|
0.525
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr19_+_49838700
|
0.525
|
|
CD37
|
CD37 molecule
|
|
chr3_-_71353910
|
0.503
|
NM_001244812
|
FOXP1
|
forkhead box P1
|
|
chr11_-_104839263
|
0.498
|
NM_001225
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase
|
|
chrX_-_154563735
|
0.496
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr6_-_33281790
|
0.493
|
|
TAPBP
|
TAP binding protein (tapasin)
|
|
chr2_-_202508223
|
0.488
|
NM_001044385
|
TMEM237
|
transmembrane protein 237
|
|
chr5_+_6746100
|
0.482
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr12_-_31479084
|
0.465
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr1_+_111889194
|
0.462
|
NM_181643
|
C1orf88
|
chromosome 1 open reading frame 88
|
|
chr11_-_5706186
|
0.461
|
NM_033034
NM_033092
NM_033093
|
TRIM5
|
tripartite motif containing 5
|
|
chr1_+_200863948
|
0.450
|
NM_001142569
|
C1orf106
|
chromosome 1 open reading frame 106
|
|
chr6_-_32812189
|
0.448
|
|
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7)
|
|
chr12_-_56709831
|
0.441
|
NM_001190991
NM_014255
|
CNPY2
|
canopy 2 homolog (zebrafish)
|
|
chr3_-_11623829
|
0.440
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr2_+_169643048
|
0.436
|
NM_001171631
|
NOSTRIN
|
nitric oxide synthase trafficker
|
|
chr6_-_130031278
|
0.429
|
NM_033515
|
ARHGAP18
|
Rho GTPase activating protein 18
|
|
chr19_+_49838734
|
0.426
|
|
CD37
|
CD37 molecule
|
|
chr15_+_71184781
|
0.423
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr19_-_11639929
|
0.420
|
NM_001142464
NM_001142465
NM_016581
|
ECSIT
|
ECSIT homolog (Drosophila)
|
|
chr11_-_104905849
|
0.417
|
|
CASP1
|
caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase)
|
|
chr12_-_9913415
|
0.405
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr14_-_24701559
|
0.399
|
|
NEDD8
|
neural precursor cell expressed, developmentally down-regulated 8
|
|
chr6_+_25962978
|
0.398
|
NM_006355
|
TRIM38
|
tripartite motif containing 38
|
|
chr11_+_64008530
|
0.398
|
NM_001135208
|
FKBP2
|
FK506 binding protein 2, 13kDa
|
|
chr3_+_51976274
|
0.394
|
NM_001003931
NM_005485
|
PARP3
|
poly (ADP-ribose) polymerase family, member 3
|
|
chr7_+_120702818
|
0.392
|
|
C7orf58
|
chromosome 7 open reading frame 58
|
|
chr6_+_3259185
|
0.386
|
|
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4
|
|
chr11_+_77532159
|
0.378
|
NM_024684
|
C11orf67
|
chromosome 11 open reading frame 67
|
|
chr10_+_5238756
|
0.377
|
NM_001818
|
AKR1C4
|
aldo-keto reductase family 1, member C4 (chlordecone reductase; 3-alpha hydroxysteroid dehydrogenase, type I; dihydrodiol dehydrogenase 4)
|
|
chr22_-_36562224
|
0.372
|
NM_145641
NM_145642
|
APOL3
|
apolipoprotein L, 3
|
|
chr1_-_20445991
|
0.372
|
NM_012400
|
PLA2G2D
|
phospholipase A2, group IID
|
|
chr11_-_128392061
|
0.372
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr19_-_42192095
|
0.367
|
NM_006890
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7
|
|
chr1_+_226012974
|
0.363
|
NM_000120
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic)
|
|
chr12_-_31479038
|
0.363
|
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr17_-_80407601
|
0.359
|
NM_001193655
|
C17orf62
|
chromosome 17 open reading frame 62
|
|
chr1_-_144520918
|
0.358
|
|
LOC728855
|
uncharacterized LOC728855
|
|
chr9_+_33624214
|
0.342
|
|
ANXA2P2
|
annexin A2 pseudogene 2
|
|
chr12_-_31479120
|
0.338
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr19_-_18197632
|
0.318
|
NM_005535
NM_153701
|
IL12RB1
|
interleukin 12 receptor, beta 1
|