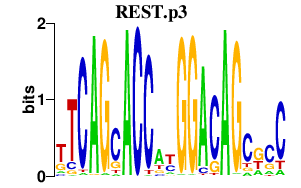
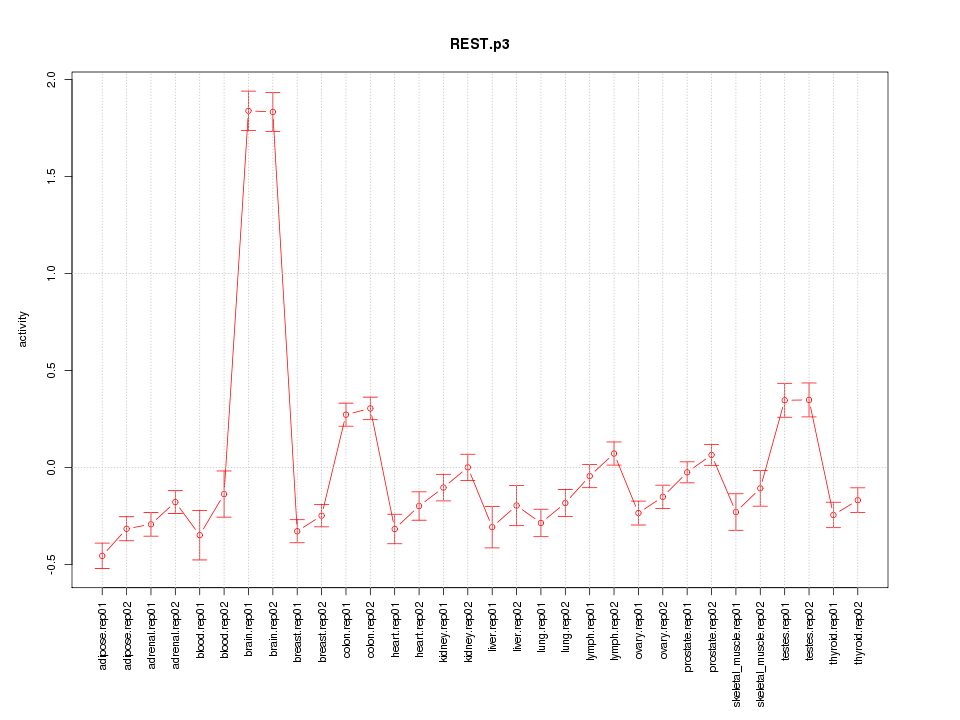
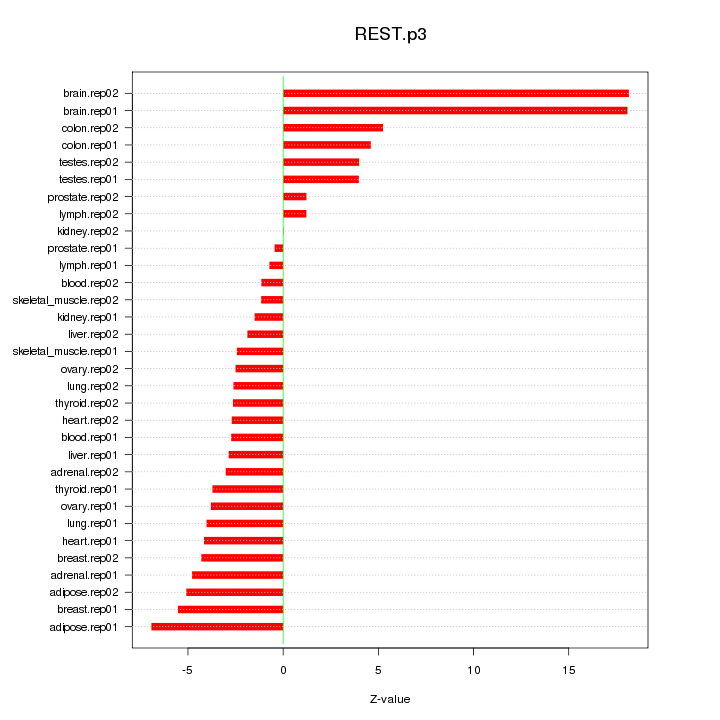
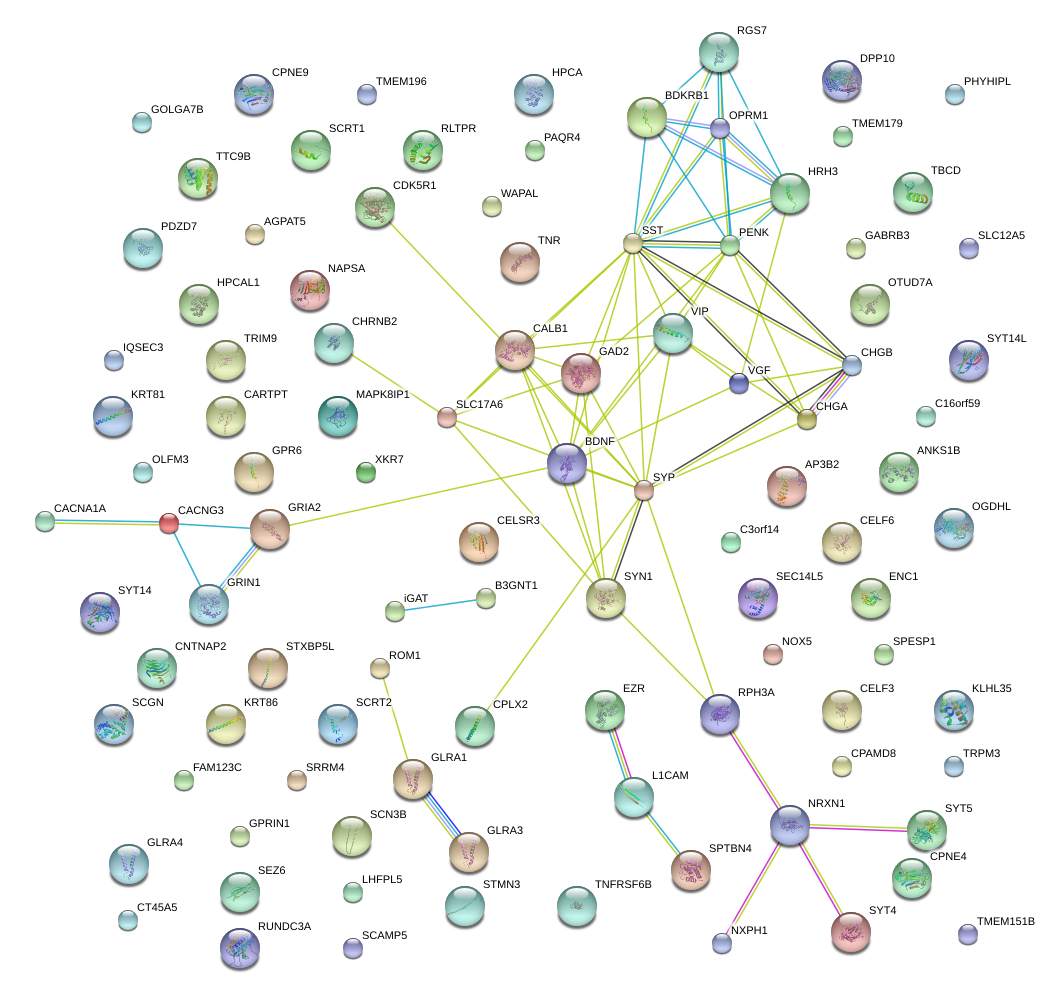
|
chr5_+_71014989
|
67.768
|
NM_004291
|
CARTPT
|
CART prepropeptide
|
|
chr14_+_93389491
|
42.791
|
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr1_+_33352071
|
42.432
|
NM_002143
|
HPCA
|
hippocalcin
|
|
chr17_+_42385780
|
37.778
|
NM_001144825
NM_001144826
NM_006695
|
RUNDC3A
|
RUN domain containing 3A
|
|
chr20_+_5891973
|
37.715
|
NM_001819
|
CHGB
|
chromogranin B (secretogranin 1)
|
|
chr9_+_140033588
|
36.522
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr20_+_44657858
|
34.995
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr19_-_55691468
|
34.604
|
NM_003180
|
SYT5
|
synaptotagmin V
|
|
chrX_-_49056638
|
33.178
|
NM_003179
|
SYP
|
synaptophysin
|
|
chr5_+_175298492
|
32.642
|
NM_001008220
|
CPLX2
|
complexin 2
|
|
chr4_+_158141735
|
32.082
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr3_-_187388105
|
31.425
|
NM_001048
|
SST
|
somatostatin
|
|
chr1_-_241520467
|
30.894
|
NM_002924
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr12_+_113229548
|
30.285
|
NM_001143854
NM_014954
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr10_+_26505593
|
30.080
|
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr1_-_102462371
|
29.655
|
|
OLFM3
|
olfactomedin 3
|
|
chr1_+_154540250
|
28.858
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr12_+_113229713
|
28.473
|
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr16_+_24267672
|
28.341
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chrX_-_153141284
|
28.099
|
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr15_-_83378488
|
27.982
|
NM_004644
|
AP3B2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr5_+_175298538
|
27.926
|
|
CPLX2
|
complexin 2
|
|
chr14_+_93389444
|
27.609
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr14_-_51561657
|
25.984
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr19_+_4769144
|
25.778
|
|
MIR7-3HG
|
MIR7-3 host gene (non-protein coding)
|
|
chr7_-_100808832
|
25.650
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr20_+_44657812
|
25.505
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr5_+_175298604
|
25.087
|
|
CPLX2
|
complexin 2
|
|
chr7_+_8474016
|
25.083
|
|
NXPH1
|
neurexophilin 1
|
|
chrX_-_153141398
|
24.425
|
NM_000425
NM_001143963
NM_024003
|
L1CAM
|
L1 cell adhesion molecule
|
|
chr8_-_91094924
|
24.224
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr20_-_10200137
|
23.887
|
|
LOC100131208
|
uncharacterized LOC100131208
|
|
chr3_+_120627026
|
23.868
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr3_-_131753757
|
23.695
|
NM_130808
|
CPNE4
|
copine IV
|
|
chr16_+_24267474
|
23.418
|
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3
|
|
chr12_+_119419299
|
23.191
|
NM_194286
|
SRRM4
|
serine/arginine repetitive matrix 4
|
|
chr19_-_40724272
|
22.654
|
NM_152479
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr8_-_145559831
|
22.610
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr10_+_60937226
|
22.585
|
NM_001143774
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr20_-_60795270
|
22.228
|
NM_007232
|
HRH3
|
histamine receptor H3
|
|
chr10_+_26505235
|
22.088
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr10_-_50970321
|
21.815
|
NM_001143996
NM_001143997
NM_018245
|
OGDHL
|
oxoglutarate dehydrogenase-like
|
|
chr2_-_51259536
|
21.278
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr11_+_22359666
|
20.882
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr5_-_176037091
|
20.737
|
NM_052899
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr1_+_210111537
|
20.675
|
NM_001146261
NM_001146262
NM_001146264
NM_153262
|
SYT14
|
synaptotagmin XIV
|
|
chr11_+_45907198
|
20.631
|
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr3_+_62304667
|
20.602
|
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chrX_-_47479095
|
20.340
|
|
|
|
|
chr2_+_115199898
|
20.031
|
NM_020868
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr1_+_154540300
|
19.988
|
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr7_-_19812403
|
19.853
|
NM_152774
|
TMEM196
|
transmembrane protein 196
|
|
chr7_+_8473584
|
19.655
|
NM_152745
|
NXPH1
|
neurexophilin 1
|
|
chr8_-_57358511
|
19.582
|
NM_006211
|
PENK
|
proenkephalin
|
|
chr3_+_62305395
|
19.430
|
NM_020685
|
C3orf14
|
chromosome 3 open reading frame 14
|
|
chrX_-_47479229
|
19.329
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr14_-_51562044
|
19.290
|
|
TRIM9
|
tripartite motif containing 9
|
|
chr1_-_175712669
|
19.176
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr1_+_210111575
|
19.165
|
|
SYT14
|
synaptotagmin XIV
|
|
chr17_+_30813928
|
19.126
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr11_-_123524881
|
19.025
|
|
SCN3B
|
sodium channel, voltage-gated, type III, beta
|
|
chr11_+_45907046
|
18.628
|
NM_005456
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr6_+_110300197
|
18.571
|
NM_005284
|
GPR6
|
G protein-coupled receptor 6
|
|
chr14_-_105071080
|
18.171
|
NM_207379
|
TMEM179
|
transmembrane protein 179
|
|
chr10_+_99609994
|
17.749
|
NM_001010917
|
GOLGA7B
|
golgin A7 family, member B
|
|
chr18_-_40857245
|
17.631
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr3_+_9745509
|
17.601
|
NM_153635
|
CPNE9
|
copine family member IX
|
|
chr15_-_27018917
|
17.193
|
NM_021912
|
GABRB3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3
|
|
chr17_-_27332888
|
17.105
|
NM_001098635
NM_178860
|
SEZ6
|
seizure related 6 homolog (mouse)
|
|
chr1_-_102462789
|
16.885
|
NM_058170
|
OLFM3
|
olfactomedin 3
|
|
chr19_+_41036430
|
16.653
|
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr14_-_74892804
|
16.553
|
NM_001105579
|
SYNDIG1L
|
synapse differentiation inducing 1-like
|
|
chr16_+_5008317
|
15.687
|
NM_014692
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae)
|
|
chr15_+_75287829
|
15.462
|
NM_001178111
NM_001178112
NM_138967
|
SCAMP5
|
secretory carrier membrane protein 5
|
|
chr7_+_8473237
|
15.204
|
|
NXPH1
|
neurexophilin 1
|
|
chr1_-_151689246
|
15.024
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr19_+_41036367
|
14.373
|
NM_025213
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr1_-_241520384
|
13.368
|
|
RGS7
|
regulator of G-protein signaling 7
|
|
chr18_-_5197240
|
13.248
|
NM_001145194
|
C18orf42
|
chromosome 18 open reading frame 42
|
|
chr7_+_145813452
|
13.155
|
NM_014141
|
CNTNAP2
|
contactin associated protein-like 2
|
|
chr12_+_175872
|
13.056
|
NM_001170738
|
IQSEC3
|
IQ motif and Sec7 domain 3
|
|
chr6_+_35773070
|
13.036
|
NM_182548
|
LHFPL5
|
lipoma HMGIC fusion partner-like 5
|
|
chr4_-_175750377
|
12.803
|
NM_001042543
NM_006529
|
GLRA3
|
glycine receptor, alpha 3
|
|
chr11_-_75141015
|
11.082
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr10_-_102790850
|
10.511
|
|
PDZD7
|
PDZ domain containing 7
|
|
chr2_-_51259239
|
10.332
|
|
NRXN1
|
neurexin 1
|
|
chr16_+_67686813
|
10.311
|
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing
|
|
chr20_-_62284701
|
9.671
|
NM_015894
|
STMN3
|
stathmin-like 3
|
|
chr12_-_3601984
|
9.490
|
|
LOC100129223
|
uncharacterized LOC100129223
|
|
chr6_+_44238479
|
9.375
|
NM_001137560
|
TMEM151B
|
transmembrane protein 151B
|
|
chr11_-_66115016
|
9.096
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr2_+_207507141
|
8.287
|
NM_001102659
|
LOC200726
|
hCG1657980
|
|
chr6_+_153071931
|
8.282
|
NM_003381
NM_194435
|
VIP
|
vasoactive intestinal peptide
|
|
chr16_+_3019341
|
8.181
|
NM_152341
|
PAQR4
|
progestin and adipoQ receptor family member IV
|
|
chr11_+_62380212
|
8.064
|
NM_000327
|
ROM1
|
retinal outer segment membrane protein 1
|
|
chr11_-_75141210
|
7.901
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr15_-_32162778
|
7.265
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr2_+_131513782
|
6.614
|
NM_001105194
NM_001105195
|
FAM123C
|
family with sequence similarity 123C
|
|
chr19_-_39402797
|
6.589
|
NM_001243212
|
LOC643669
|
uncharacterized LOC643669
|
|
chr3_-_48700317
|
6.479
|
NM_001407
|
CELSR3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr12_+_52695648
|
5.224
|
NM_002284
|
KRT86
|
keratin 86
|
|
chr20_+_30555804
|
4.889
|
NM_001011718
|
XKR7
|
XK, Kell blood group complex subunit-related family, member 7
|
|
chr15_-_72612127
|
4.693
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr11_-_75141517
|
4.391
|
NM_001039548
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr6_+_154360442
|
4.330
|
NM_000914
NM_001008503
NM_001008504
NM_001008505
NM_001145282
NM_001145283
NM_001145284
NM_001145285
NM_001145286
|
OPRM1
|
opioid receptor, mu 1
|
|
chr9_-_74061819
|
4.257
|
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr6_+_25652373
|
4.158
|
NM_006998
|
SCGN
|
secretagogin, EF-hand calcium binding protein
|
|
chr15_-_32162896
|
3.694
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr12_-_99288502
|
2.792
|
NM_001204065
NM_001204079
NM_001204080
NM_001204081
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr10_-_102790877
|
2.695
|
NM_001195263
NM_024895
|
PDZD7
|
PDZ domain containing 7
|
|
chr8_+_57358361
|
2.608
|
|
|
|
|
chr20_+_62328002
|
2.406
|
NM_003823
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr20_+_62328068
|
2.183
|
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy
|
|
chr8_+_6565870
|
2.158
|
NM_018361
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr8_+_6566153
|
2.003
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr15_+_69222838
|
1.956
|
NM_145658
NM_001184780
|
SPESP1
NOX5
|
sperm equatorial segment protein 1
NADPH oxidase, EF-hand calcium binding domain 5
|
|
chr16_+_2510080
|
1.842
|
NM_025108
|
C16orf59
|
chromosome 16 open reading frame 59
|
|
chr8_+_6565914
|
1.678
|
|
AGPAT5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr11_-_27721179
|
1.348
|
NM_170734
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr14_-_58332591
|
1.298
|
NM_001206920
|
SLC35F4
|
solute carrier family 35, member F4
|
|
chr11_-_27742224
|
1.222
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_128807712
|
0.955
|
NM_001195194
|
TP53AIP1
|
tumor protein p53 regulated apoptosis inducing protein 1
|
|
chr12_+_71833518
|
0.907
|
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr6_-_71666740
|
0.901
|
NM_080742
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S)
|
|
chr17_+_73723729
|
0.889
|
|
ITGB4
|
integrin, beta 4
|
|
chr2_-_42720476
|
0.746
|
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr17_+_80332152
|
0.626
|
NM_018949
|
UTS2R
|
urotensin 2 receptor
|
|
chr17_-_65362667
|
0.596
|
NM_002816
NM_174871
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr17_+_1173857
|
0.539
|
NM_001164405
|
BHLHA9
|
basic helix-loop-helix family, member a9
|
|
chr8_+_68864723
|
0.471
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr8_+_68864589
|
0.419
|
NM_024870
NM_025170
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr9_-_104357202
|
0.315
|
NM_147180
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta
|
|
chr17_-_65362635
|
0.306
|
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr5_+_69696805
|
0.302
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr5_-_69881965
|
0.265
|
|
SMA4
|
glucuronidase, beta pseudogene
|
|
chr8_-_93977715
|
0.237
|
NM_001191036
|
C8orf83
|
chromosome 8 open reading frame 83
|
|
chr11_-_134123138
|
0.202
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr11_+_32112607
|
0.177
|
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain
|
|
chr12_+_6561176
|
0.158
|
NM_018009
|
TAPBPL
|
TAP binding protein-like
|
|
chr9_-_123812535
|
0.074
|
NM_001735
|
C5
|
complement component 5
|
|
chr22_-_18256748
|
0.074
|
NM_001244572
NM_197966
|
BID
|
BH3 interacting domain death agonist
|
|
chr12_+_51347781
|
0.058
|
NM_001109619
|
HIGD1C
|
HIG1 hypoxia inducible domain family, member 1C
|
|
chr1_-_33336289
|
0.033
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr1_+_101004456
|
0.004
|
|
GPR88
|
G protein-coupled receptor 88
|