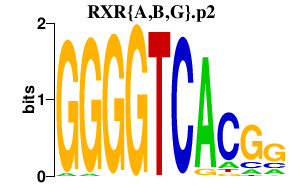
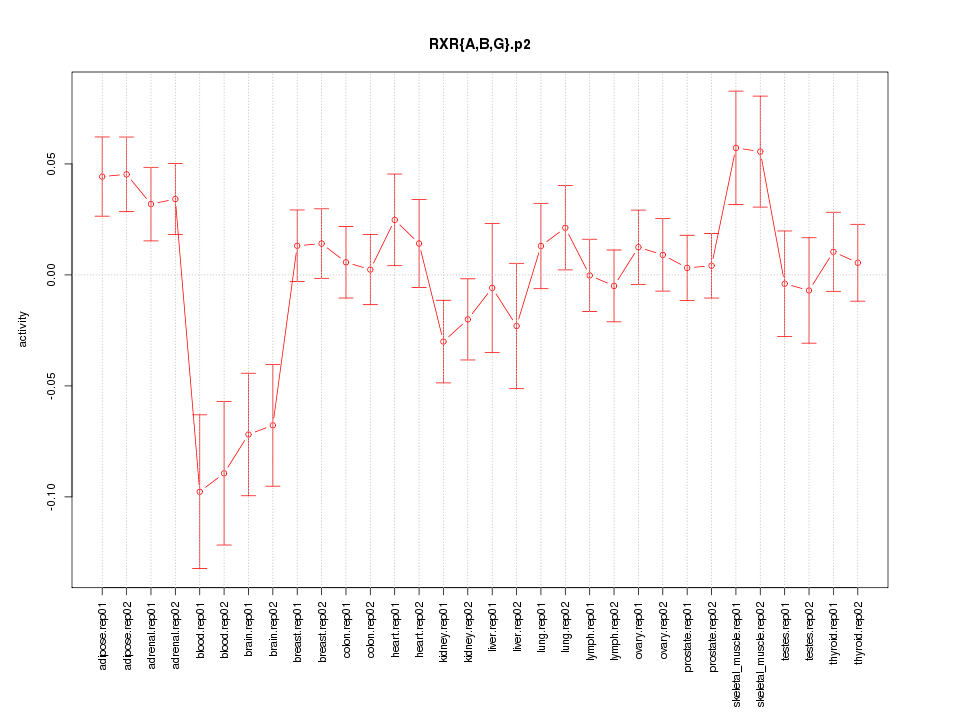
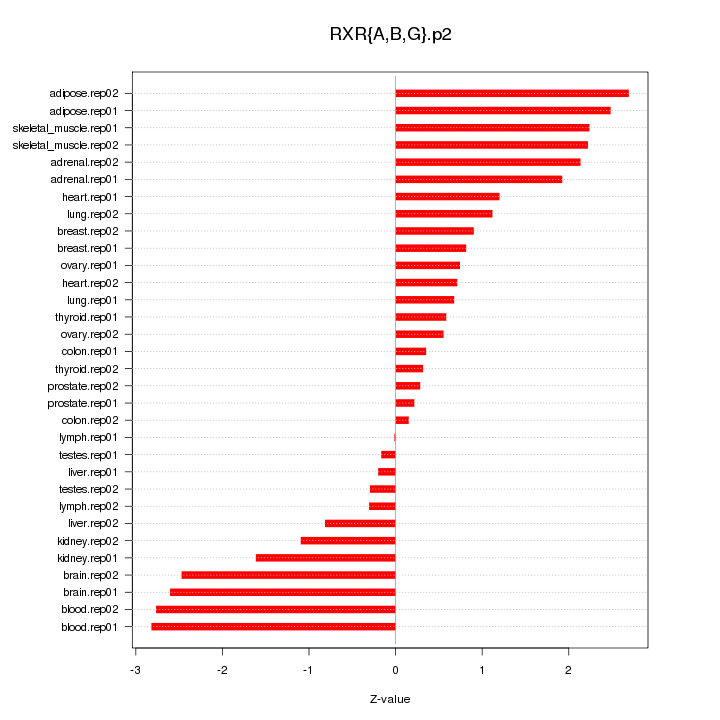
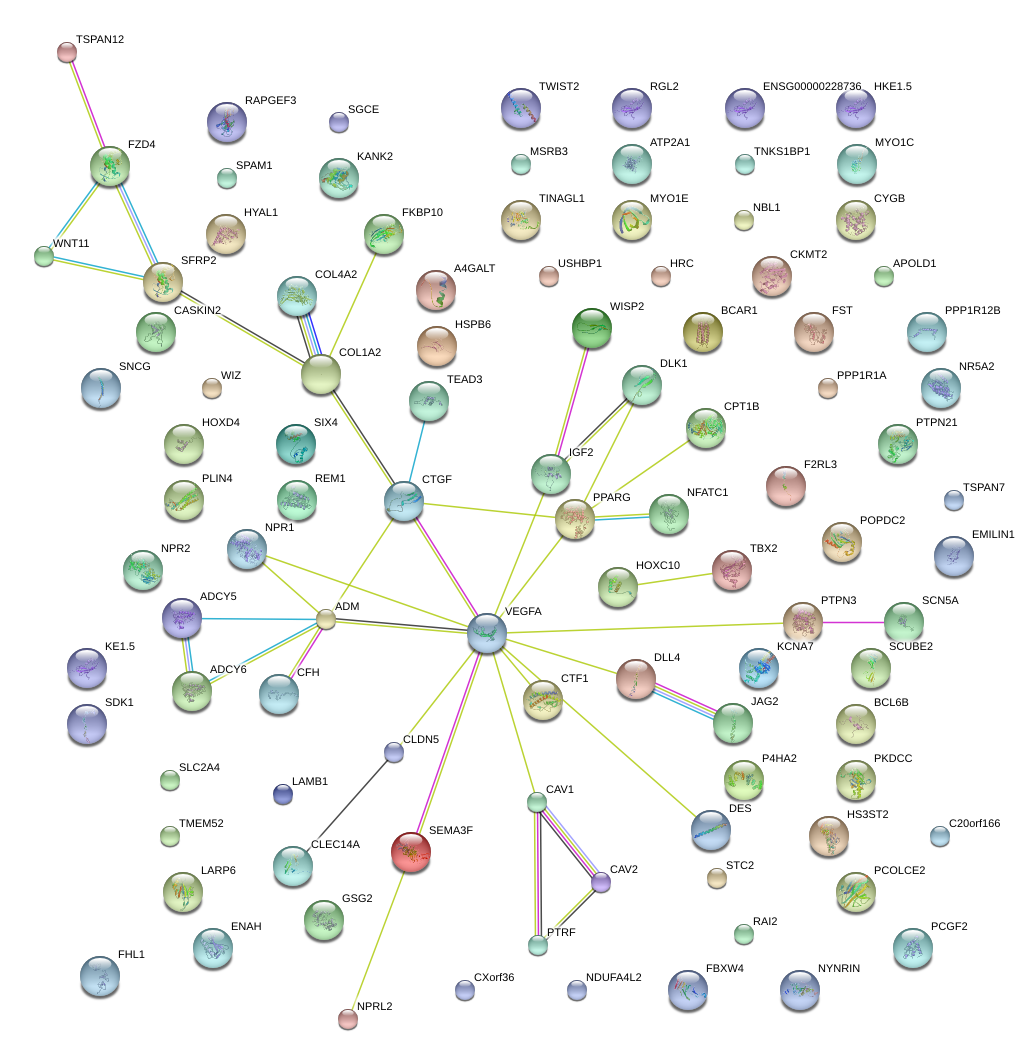
|
chr17_+_7184978
|
6.021
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr7_+_116166346
|
4.783
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr19_-_49658645
|
4.538
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr11_-_2017822
|
3.854
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr11_-_2018446
|
3.767
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr12_+_54378929
|
3.767
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr19_-_36246411
|
3.556
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr11_-_2018334
|
3.261
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr19_-_36246206
|
3.003
|
|
HSPB6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr2_+_27301693
|
2.879
|
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chr2_+_42275017
|
2.858
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr14_-_61190458
|
2.825
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr6_+_43738394
|
2.758
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr6_+_43737937
|
2.740
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr3_-_123167391
|
2.736
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr12_-_54982271
|
2.719
|
NM_006741
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr1_+_202317798
|
2.716
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B
|
|
chr15_+_41221530
|
2.713
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr5_-_172755195
|
2.685
|
|
STC2
|
stanniocalcin 2
|
|
chr19_+_16999871
|
2.634
|
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chr5_-_172755418
|
2.466
|
|
STC2
|
stanniocalcin 2
|
|
chr1_-_1850704
|
2.457
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr22_-_51016343
|
2.457
|
NM_001145137
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr5_-_131563481
|
2.439
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr20_+_43343884
|
2.426
|
NM_003881
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr14_-_38725539
|
2.355
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr14_-_38725253
|
2.343
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr11_-_86666211
|
2.335
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr6_+_43738177
|
2.309
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr17_+_6926366
|
2.302
|
NM_181844
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr11_-_86666089
|
2.221
|
|
FZD4
|
frizzled family receptor 4
|
|
chr6_+_43738757
|
2.127
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr14_+_24867926
|
2.068
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr7_+_116139619
|
2.042
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr3_+_50192847
|
2.007
|
NM_004186
|
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F
|
|
chr14_-_89020888
|
2.007
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr22_-_19511777
|
1.967
|
|
CLDN5
|
claudin 5
|
|
chr11_-_75917450
|
1.952
|
NM_004626
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr1_+_19972259
|
1.950
|
NM_001204086
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr17_-_74533623
|
1.913
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr1_+_32042125
|
1.902
|
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr2_+_220283098
|
1.831
|
NM_001927
|
DES
|
desmin
|
|
chr17_-_40575117
|
1.798
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr7_+_116140097
|
1.785
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr19_+_16999807
|
1.784
|
NM_003950
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3
|
|
chrX_-_17879355
|
1.775
|
NM_001172732
NM_001172739
NM_001172743
NM_021785
|
RAI2
|
retinoic acid induced 2
|
|
chr2_+_239756672
|
1.773
|
NM_057179
|
TWIST2
|
twist homolog 2 (Drosophila)
|
|
chr17_+_39969205
|
1.767
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr19_-_15560761
|
1.761
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr11_-_9113092
|
1.739
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr9_-_112260529
|
1.731
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr20_+_30063090
|
1.725
|
NM_014012
|
REM1
|
RAS (RAD and GEM)-like GTP-binding 1
|
|
chr17_+_6926414
|
1.707
|
|
BCL6B
|
B-cell CLL/lymphoma 6, member B
|
|
chr16_+_30907927
|
1.695
|
NM_001142544
NM_001330
|
CTF1
|
cardiotrophin 1
|
|
chr2_+_27301434
|
1.668
|
NM_007046
|
EMILIN1
|
elastin microfibril interfacer 1
|
|
chrX_+_38420690
|
1.659
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr10_+_88718287
|
1.631
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr12_-_57634474
|
1.626
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr3_-_142607737
|
1.609
|
NM_013363
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2
|
|
chr20_+_61147924
|
1.607
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr11_-_2162340
|
1.601
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr13_+_111138079
|
1.544
|
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr6_-_132272311
|
1.529
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr7_-_107643330
|
1.522
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr12_+_12938540
|
1.513
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr10_-_103454687
|
1.505
|
NM_022039
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr12_-_48152180
|
1.500
|
NM_001098532
NM_006105
|
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_50340979
|
1.490
|
NM_007312
NM_033159
NM_153282
NM_153283
NM_153285
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr19_-_4517715
|
1.477
|
NM_001080400
|
PLIN4
|
perilipin 4
|
|
chr16_-_75299713
|
1.476
|
NM_001170720
NM_001170714
NM_001170718
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr1_+_182992694
|
1.469
|
|
|
|
|
chr1_+_199996729
|
1.456
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr22_-_43116796
|
1.452
|
NM_017436
|
A4GALT
|
alpha 1,4-galactosyltransferase
|
|
chr14_-_105635114
|
1.444
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr2_+_177016112
|
1.438
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr16_+_28889808
|
1.434
|
NM_004320
NM_173201
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr6_-_33266297
|
1.420
|
|
RGL2
|
ral guanine nucleotide dissociation stimulator-like 2
|
|
chr3_-_119379269
|
1.414
|
NM_022135
|
POPDC2
|
popeye domain containing 2
|
|
chr7_-_94285477
|
1.414
|
NM_001099400
NM_001099401
NM_003919
|
SGCE
|
sarcoglycan, epsilon
|
|
chr17_+_59477169
|
1.414
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr20_+_43344005
|
1.398
|
|
WISP2
|
WNT1 inducible signaling pathway protein 2
|
|
chr3_+_12329332
|
1.390
|
NM_005037
NM_138712
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr12_+_12878718
|
1.389
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr19_-_17375522
|
1.371
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr21_+_46825291
|
1.370
|
|
|
|
|
chr16_+_22825806
|
1.358
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr7_+_94024201
|
1.356
|
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr5_-_138730884
|
1.355
|
NM_001161546
|
C5orf65
|
chromosome 5 open reading frame 65
|
|
chr11_-_57092283
|
1.350
|
NM_033396
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa
|
|
chr5_+_80529138
|
1.341
|
NM_001099735
NM_001099736
NM_001825
|
CKMT2
|
creatine kinase, mitochondrial 2 (sarcomeric)
|
|
chr18_+_77160243
|
1.330
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr17_-_1394849
|
1.318
|
NM_033375
|
MYO1C
|
myosin IC
|
|
chr22_-_19512001
|
1.314
|
|
CLDN5
|
claudin 5
|
|
chr4_-_154710207
|
1.313
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr17_+_39968961
|
1.311
|
NM_021939
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr19_-_49576120
|
1.291
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr12_+_65672422
|
1.289
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chrX_-_45060081
|
1.278
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr1_+_19971865
|
1.267
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr15_-_71146427
|
1.264
|
NM_018357
NM_197958
|
LARP6
|
La ribonucleoprotein domain family, member 6
|
|
chrX_+_135229536
|
1.256
|
NM_001159702
NM_001159703
NM_001449
|
FHL1
|
four and a half LIM domains 1
|
|
chr17_-_36906030
|
1.231
|
|
PCGF2
|
polycomb group ring finger 2
|
|
chr7_-_120498009
|
1.230
|
|
TSPAN12
|
tetraspanin 12
|
|
chr9_+_35792405
|
1.228
|
NM_003995
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B)
|
|
chr7_+_3340919
|
1.206
|
NM_152744
|
SDK1
|
sidekick cell adhesion molecule 1
|
|
chr11_-_2018703
|
1.203
|
|
H19
|
H19, imprinted maternally expressed transcript (non-protein coding)
|
|
chr6_-_35464716
|
1.195
|
NM_003214
|
TEAD3
|
TEA domain family member 3
|
|
chr14_+_101193190
|
1.192
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr1_-_225840660
|
1.178
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr17_+_3627133
|
1.178
|
NM_031965
|
GSG2
|
germ cell associated 2 (haspin)
|
|
chr17_-_73511626
|
1.174
|
NM_020753
|
CASKIN2
|
CASK interacting protein 2
|
|
chr19_-_11308184
|
1.172
|
NM_001136191
|
KANK2
|
KN motif and ankyrin repeat domains 2
|
|
chr5_+_52776263
|
1.171
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr3_-_38691118
|
1.170
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr11_+_10326618
|
1.169
|
NM_001124
|
ADM
|
adrenomedullin
|
|
chr7_-_27239724
|
1.169
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr17_+_1958354
|
1.167
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr7_-_94285426
|
1.167
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr7_-_94285372
|
1.154
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr20_+_53092131
|
1.152
|
NM_018431
|
DOK5
|
docking protein 5
|
|
chr17_-_73511453
|
1.140
|
|
CASKIN2
|
CASK interacting protein 2
|
|
chr1_-_154946830
|
1.139
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr7_+_94023872
|
1.139
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr14_-_105634708
|
1.129
|
|
JAG2
|
jagged 2
|
|
chr17_+_39969200
|
1.128
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr11_-_11643542
|
1.125
|
NM_198516
|
GALNTL4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chr9_-_110251754
|
1.122
|
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chrX_-_34675366
|
1.118
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr7_+_75932992
|
1.118
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr17_+_41003360
|
1.109
|
|
|
|
|
chr11_-_63933204
|
1.104
|
|
MACROD1
|
MACRO domain containing 1
|
|
chr13_+_110959592
|
1.103
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr5_+_131593388
|
1.101
|
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr16_-_73092533
|
1.101
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr10_-_103454909
|
1.094
|
|
FBXW4
|
F-box and WD repeat domain containing 4
|
|
chr2_+_220309373
|
1.094
|
|
SPEG
|
SPEG complex locus
|
|
chr19_-_56056908
|
1.089
|
NM_001199824
|
SGK110
|
putative uncharacterized serine/threonine-protein kinase SgK110-like
|
|
chr17_-_79792867
|
1.087
|
|
PPP1R27
|
protein phosphatase 1, regulatory subunit 27
|
|
chr15_+_62682682
|
1.081
|
|
TLN2
|
talin 2
|
|
chr7_-_82073020
|
1.080
|
NM_000722
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chrX_-_132548994
|
1.080
|
NM_001448
|
GPC4
|
glypican 4
|
|
chr1_+_32042085
|
1.078
|
NM_001204414
NM_022164
|
TINAGL1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr11_-_47447915
|
1.071
|
NM_002804
|
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3
|
|
chr12_+_65672682
|
1.069
|
NM_001193461
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr3_-_50329702
|
1.063
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr7_-_82072804
|
1.060
|
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1
|
|
chr1_-_154946801
|
1.059
|
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr9_+_137533643
|
1.057
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr13_-_76055872
|
1.056
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr19_+_55999921
|
1.048
|
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains)
|
|
chr13_+_114462148
|
1.045
|
NM_182614
|
FAM70B
|
family with sequence similarity 70, member B
|
|
chr11_-_65640199
|
1.044
|
NM_016938
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2
|
|
chrX_-_132549421
|
1.042
|
|
GPC4
|
glypican 4
|
|
chr14_-_89883306
|
1.039
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr11_-_65430545
|
1.037
|
|
|
|
|
chr10_+_81892257
|
1.034
|
NM_001012973
|
PLAC9
|
placenta-specific 9
|
|
chr20_-_36793646
|
1.034
|
NM_004613
NM_198951
|
TGM2
|
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase)
|
|
chr3_-_50360109
|
1.032
|
NM_003773
NM_033158
|
HYAL2
|
hyaluronoglucosaminidase 2
|
|
chr19_-_4723814
|
1.029
|
NM_139159
|
DPP9
|
dipeptidyl-peptidase 9
|
|
chr6_+_31973358
|
1.026
|
NM_000500
NM_001128590
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2
|
|
chr17_+_59477473
|
1.024
|
|
TBX2
|
T-box 2
|
|
chr19_-_6424820
|
1.022
|
NM_003685
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr11_+_450346
|
1.021
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr16_+_55358470
|
1.021
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr14_+_23305913
|
1.015
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr8_+_145321462
|
1.010
|
NM_001008271
NM_001080514
|
SCXA
SCXB
|
scleraxis homolog A (mouse)
scleraxis homolog B (mouse)
|
|
chr5_-_134369571
|
1.009
|
|
PITX1
|
paired-like homeodomain 1
|
|
chr12_-_57630867
|
1.009
|
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr9_-_124990949
|
1.009
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr1_-_154946859
|
1.008
|
NM_001130041
NM_001202859
NM_003029
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1
|
|
chr13_-_40177304
|
1.008
|
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr11_-_61684981
|
1.006
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr17_+_12569364
|
1.006
|
|
MYOCD
|
myocardin
|
|
chr2_+_56411123
|
1.004
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr2_+_23608297
|
1.002
|
NM_052920
|
KLHL29
|
kelch-like 29 (Drosophila)
|
|
chr13_-_40177355
|
1.000
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr17_-_2614926
|
0.991
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr14_+_69726680
|
0.990
|
NM_001168368
NM_020692
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr1_-_228135587
|
0.986
|
NM_003395
|
WNT9A
|
wingless-type MMTV integration site family, member 9A
|
|
chr19_-_1174201
|
0.982
|
|
|
|
|
chr21_+_46825001
|
0.979
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr19_+_15218141
|
0.977
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr12_-_102874340
|
0.976
|
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr17_+_58227330
|
0.967
|
|
CA4
|
carbonic anhydrase IV
|
|
chr11_-_74022609
|
0.964
|
NM_182904
|
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III
|
|
chr1_-_149982528
|
0.962
|
NM_020205
|
OTUD7B
|
OTU domain containing 7B
|
|
chr15_-_55880508
|
0.960
|
|
PYGO1
|
pygopus homolog 1 (Drosophila)
|
|
chr1_-_59248287
|
0.959
|
|
JUN
|
jun proto-oncogene
|
|
chr14_+_69726868
|
0.954
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr6_-_46459010
|
0.951
|
NM_001251974
|
RCAN2
|
regulator of calcineurin 2
|
|
chr19_-_460995
|
0.946
|
NM_012435
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2
|
|
chr14_+_85996454
|
0.943
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr12_+_52400725
|
0.937
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr17_+_12569206
|
0.932
|
NM_001146312
NM_153604
|
MYOCD
|
myocardin
|
|
chr17_-_46688279
|
0.930
|
|
HOXB7
|
homeobox B7
|
|
chr17_+_7210841
|
0.927
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr14_+_101193275
|
0.926
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr17_-_38256905
|
0.919
|
NM_021724
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1
|
|
chr5_+_131593331
|
0.917
|
NM_001131027
NM_003687
|
PDLIM4
|
PDZ and LIM domain 4
|
|
chr17_+_58227278
|
0.916
|
NM_000717
|
CA4
|
carbonic anhydrase IV
|