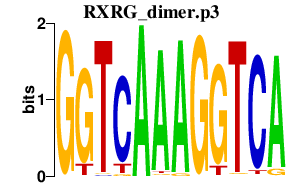
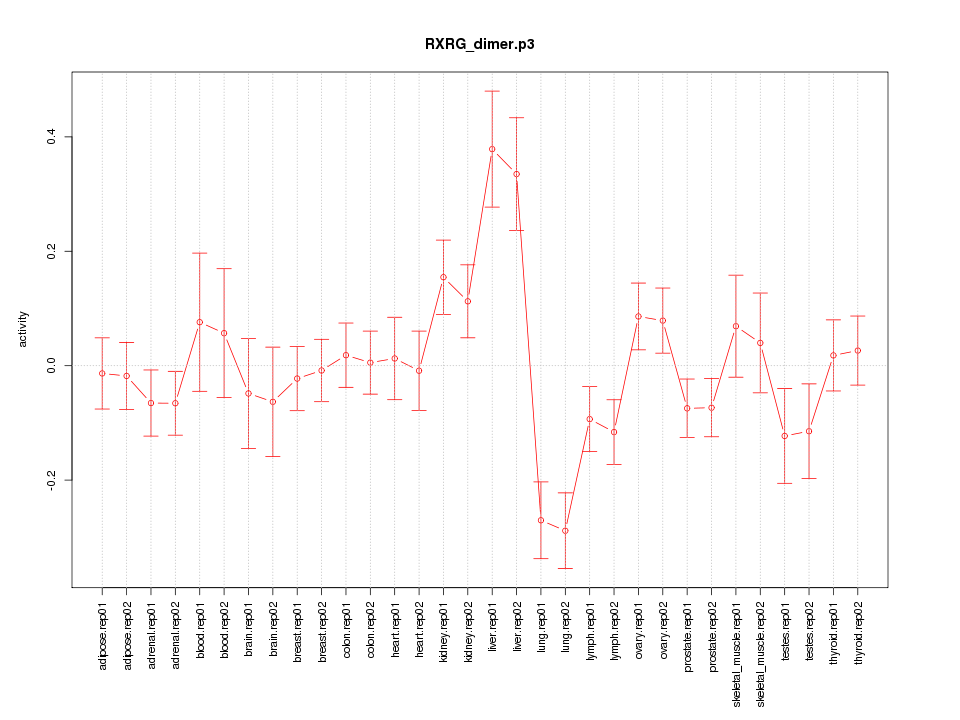
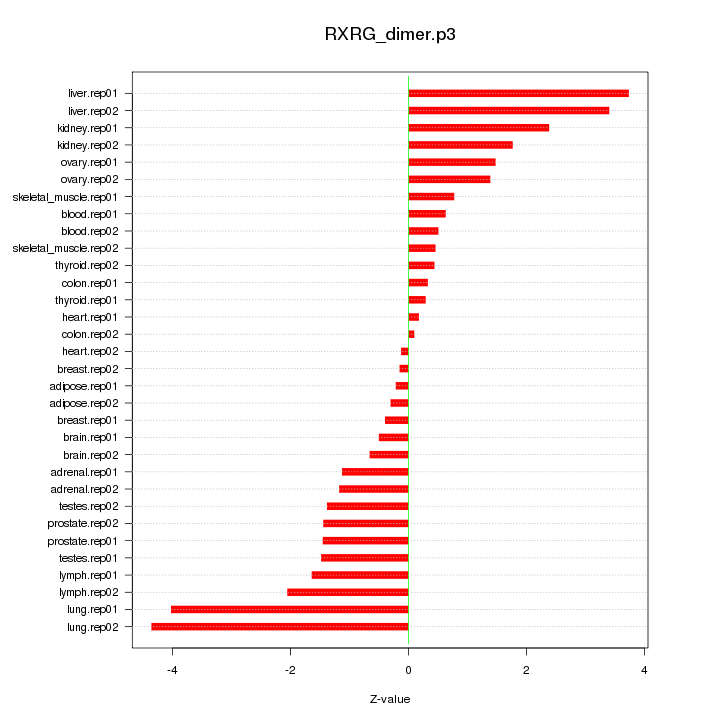
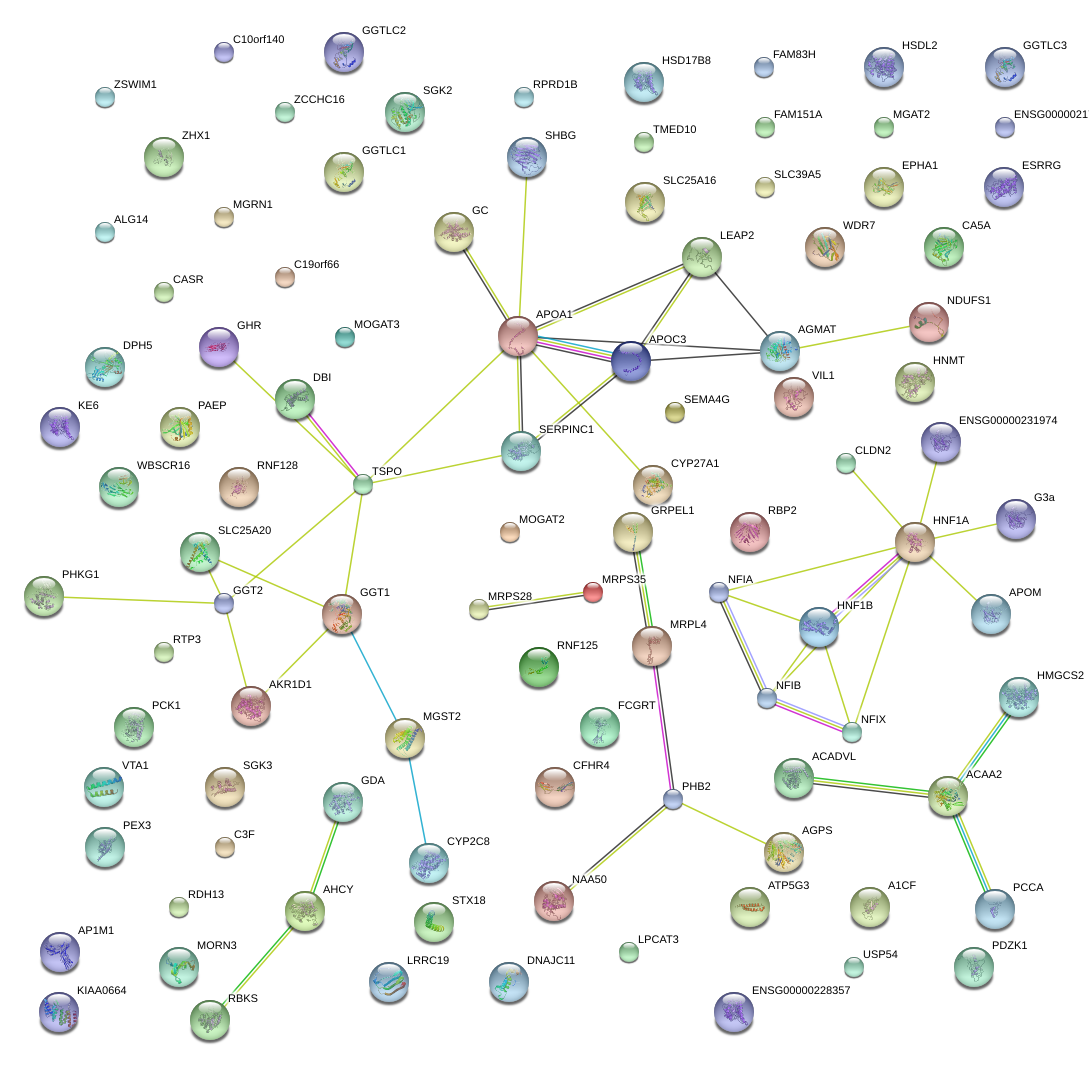
|
chr11_+_116700620
|
14.878
|
NM_000040
|
APOC3
|
apolipoprotein C-III
|
|
chr20_+_56136136
|
9.758
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr11_-_116708334
|
8.788
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr3_+_46539484
|
7.767
|
NM_031440
|
RTP3
|
receptor (chemosensory) transporter protein 3
|
|
chr4_-_72671236
|
7.186
|
NM_001204306
|
GC
|
group-specific component (vitamin D binding protein)
|
|
chr1_-_173886464
|
6.734
|
NM_000488
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr6_+_31623688
|
6.290
|
|
APOM
|
apolipoprotein M
|
|
chr19_+_45116921
|
6.080
|
NM_001205280
|
IGSF23
|
immunoglobulin superfamily, member 23
|
|
chr1_-_120311466
|
5.921
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr2_+_219283809
|
5.638
|
NM_007127
|
VIL1
|
villin 1
|
|
chr6_+_31623668
|
5.633
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr1_+_145727712
|
5.449
|
|
PDZK1
|
PDZ domain containing 1
|
|
chr10_-_96829074
|
5.314
|
NM_000770
NM_001198853
NM_001198854
NM_001198855
|
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8
|
|
chr9_+_74764266
|
5.292
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr1_+_145727665
|
5.249
|
NM_001201325
NM_001201326
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr9_+_74764394
|
4.537
|
|
GDA
|
guanine deaminase
|
|
chr10_-_52645430
|
4.502
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr10_+_102732285
|
4.414
|
NM_001203244
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr12_+_56624500
|
4.398
|
NM_001135195
|
SLC39A5
|
solute carrier family 39 (metal ion transporter), member 5
|
|
chr3_-_48936227
|
4.263
|
NM_000387
|
SLC25A20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20
|
|
chr1_-_173886401
|
4.144
|
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr9_+_74764382
|
3.816
|
|
GDA
|
guanine deaminase
|
|
chr1_-_15911461
|
3.366
|
NM_024758
|
AGMAT
|
agmatine ureohydrolase (agmatinase)
|
|
chr10_-_52645384
|
3.256
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr17_+_7533452
|
3.134
|
NM_001040
NM_001146279
NM_001146280
NM_001146281
|
SHBG
|
sex hormone-binding globulin
|
|
chr20_+_42187705
|
3.093
|
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr13_+_100741268
|
3.022
|
NM_000282
NM_001127692
NM_001178004
|
PCCA
|
propionyl CoA carboxylase, alpha polypeptide
|
|
chr17_+_7533403
|
2.930
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr9_+_74764354
|
2.869
|
|
GDA
|
guanine deaminase
|
|
chr20_+_36661849
|
2.837
|
NM_021215
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr9_+_74764417
|
2.568
|
|
GDA
|
guanine deaminase
|
|
chr9_-_27005649
|
2.549
|
NM_022901
|
LRRC19
|
leucine rich repeat containing 19
|
|
chr9_+_115142395
|
2.537
|
|
HSDL2
|
hydroxysteroid dehydrogenase like 2
|
|
chr2_-_28113211
|
2.537
|
NM_022128
|
RBKS
|
ribokinase
|
|
chr6_+_143771779
|
2.531
|
NM_003630
|
PEX3
|
peroxisomal biogenesis factor 3
|
|
chr9_+_115142188
|
2.508
|
NM_001195822
NM_032303
|
HSDL2
|
hydroxysteroid dehydrogenase like 2
|
|
chr2_-_207024103
|
2.505
|
NM_001199981
NM_001199982
NM_001199983
NM_005006
|
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase)
|
|
chr20_+_36662138
|
2.480
|
|
RPRD1B
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr17_-_2614926
|
2.409
|
NM_015229
|
KIAA0664
|
KIAA0664
|
|
chr17_-_36104874
|
2.329
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_61547625
|
2.323
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr12_+_121416294
|
2.299
|
|
HNF1A
|
HNF1 homeobox A
|
|
chrX_+_111326252
|
2.263
|
NM_001004308
|
ZCCHC16
|
zinc finger, CCHC domain containing 16
|
|
chr8_-_80942488
|
2.231
|
NM_014018
|
MRPS28
|
mitochondrial ribosomal protein S28
|
|
chr17_-_36105005
|
2.216
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr1_+_196857143
|
2.204
|
NM_001201550
NM_001201551
NM_006684
|
CFHR4
|
complement factor H-related 4
|
|
chr7_+_137761177
|
1.913
|
NM_001190906
NM_001190907
NM_005989
|
AKR1D1
|
aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase)
|
|
chr10_-_21814610
|
1.827
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr8_-_144815888
|
1.789
|
NM_198488
|
FAM83H
|
family with sequence similarity 83, member H
|
|
chr16_-_87970107
|
1.788
|
NM_001739
|
CA5A
|
carbonic anhydrase VA, mitochondrial
|
|
chr17_+_7123246
|
1.778
|
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr12_-_7125727
|
1.747
|
NM_005768
|
LPCAT3
|
lysophosphatidylcholine acyltransferase 3
|
|
chr7_-_74489572
|
1.666
|
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr1_-_55089068
|
1.663
|
NM_176782
|
FAM151A
|
family with sequence similarity 151, member A
|
|
chr1_-_217250230
|
1.650
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr4_+_140587086
|
1.634
|
|
MGST2
|
microsomal glutathione S-transferase 2
|
|
chr2_+_120125141
|
1.611
|
NM_001079863
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein)
|
|
chr7_-_100844192
|
1.587
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr20_+_36662207
|
1.547
|
|
|
|
|
chr2_-_176046120
|
1.538
|
NM_001002258
NM_001190329
NM_001689
|
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9)
|
|
chr1_+_101491401
|
1.538
|
|
|
|
|
chr19_+_10362912
|
1.522
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr3_-_113464640
|
1.459
|
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr2_+_178257371
|
1.458
|
NM_003659
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr8_-_124286489
|
1.450
|
|
ZHX1
|
zinc fingers and homeoboxes 1
|
|
chrX_+_106171328
|
1.449
|
|
CLDN2
|
claudin 2
|
|
chr1_-_6761839
|
1.429
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr1_-_6761878
|
1.397
|
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr7_-_143105897
|
1.381
|
NM_005232
|
EPHA1
|
EPH receptor A1
|
|
chr7_-_74489686
|
1.368
|
NM_030798
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chrX_+_105937067
|
1.337
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr2_+_178257488
|
1.287
|
|
AGPS
|
alkylglycerone phosphate synthase
|
|
chr1_-_95538443
|
1.246
|
NM_144988
|
ALG14
|
asparagine-linked glycosylation 14 homolog (S. cerevisiae)
|
|
chr6_+_33172418
|
1.239
|
NM_014234
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8
|
|
chr7_-_74489636
|
1.207
|
|
WBSCR16
|
Williams-Beuren syndrome chromosome region 16
|
|
chr12_+_121416548
|
1.175
|
NM_000545
|
HNF1A
|
HNF1 homeobox A
|
|
chr19_+_50016490
|
1.174
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr1_-_101491308
|
1.163
|
NM_001077394
NM_001077395
NM_015958
|
DPH5
|
DPH5 homolog (S. cerevisiae)
|
|
chr19_+_10196972
|
1.149
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr4_-_4543690
|
1.140
|
|
STX18
|
syntaxin 18
|
|
chr20_+_42187634
|
1.123
|
NM_170693
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr18_+_54318561
|
1.114
|
NM_015285
NM_052834
|
WDR7
|
WD repeat domain 7
|
|
chr20_+_44509847
|
1.113
|
NM_080603
|
ZSWIM1
|
zinc finger, SWIM-type containing 1
|
|
chr5_+_132209354
|
1.104
|
NM_052971
|
LEAP2
|
liver expressed antimicrobial peptide 2
|
|
chr18_+_29598444
|
1.086
|
NM_017831
|
RNF125
|
ring finger protein 125
|
|
chr14_+_50087400
|
1.081
|
NM_002408
|
MGAT2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase
|
|
chr2_+_219646448
|
1.079
|
NM_000784
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1
|
|
chr19_+_10362886
|
1.046
|
|
MRPL4
|
mitochondrial ribosomal protein L4
|
|
chr5_+_42548314
|
1.028
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr20_-_32899607
|
1.020
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr12_-_7079616
|
1.013
|
|
PHB2
|
prohibitin 2
|
|
chr4_-_7069755
|
1.005
|
NM_025196
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr19_+_16308664
|
0.991
|
NM_001130524
NM_032493
|
AP1M1
|
adaptor-related protein complex 1, mu 1 subunit
|
|
chr16_+_4714709
|
0.977
|
|
MGRN1
|
mahogunin, ring finger 1
|
|
chrX_+_106143393
|
0.968
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr19_-_55580896
|
0.967
|
NM_138412
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis)
|
|
chr1_-_6761897
|
0.927
|
NM_018198
|
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr18_-_47340208
|
0.926
|
NM_006111
|
ACAA2
|
acetyl-CoA acyltransferase 2
|
|
chr22_+_25003610
|
0.924
|
NM_001032365
|
GGT1
|
gamma-glutamyltransferase 1
|
|
chr6_+_142468382
|
0.914
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr14_-_75643309
|
0.909
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr14_-_75643286
|
0.908
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr2_+_138721807
|
0.902
|
NM_001024074
NM_001024075
NM_006895
|
HNMT
|
histamine N-methyltransferase
|
|
chr19_+_50016435
|
0.901
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr8_-_82607206
|
0.897
|
NM_001010893
|
SLC10A5
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 5
|
|
chr14_-_75643333
|
0.894
|
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr16_+_4475804
|
0.884
|
NM_001135110
NM_005147
|
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3
|
|
chr2_-_198364997
|
0.880
|
NM_199440
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr19_+_10197023
|
0.862
|
|
C19orf66
|
chromosome 19 open reading frame 66
|
|
chr1_-_151138330
|
0.860
|
NM_212551
NM_001136543
|
LYSMD1
|
LysM, putative peptidoglycan-binding, domain containing 1
|
|
chr20_-_20033005
|
0.846
|
|
CRNKL1
|
crooked neck pre-mRNA splicing factor-like 1 (Drosophila)
|
|
chr18_-_47339854
|
0.839
|
|
ACAA2
|
acetyl-CoA acyltransferase 2
|
|
chr11_-_45939361
|
0.821
|
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr6_+_142468415
|
0.813
|
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr10_-_129691047
|
0.796
|
|
|
|
|
chr7_+_97841565
|
0.778
|
NM_177455
|
BHLHA15
|
basic helix-loop-helix family, member a15
|
|
chr10_+_695887
|
0.764
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr7_-_65447187
|
0.751
|
|
GUSB
|
glucuronidase, beta
|
|
chr18_+_29598787
|
0.751
|
|
RNF125
|
ring finger protein 125
|
|
chr12_+_50135953
|
0.750
|
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6
|
|
chr6_+_142468401
|
0.732
|
NM_016485
|
VTA1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr14_-_75643335
|
0.730
|
NM_006827
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr19_+_50016500
|
0.719
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_+_100318422
|
0.708
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr22_+_30163351
|
0.703
|
NM_001003684
NM_013387
|
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr1_-_40042461
|
0.699
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr5_+_14664803
|
0.695
|
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr22_+_41865112
|
0.685
|
NM_001098
|
ACO2
|
aconitase 2, mitochondrial
|
|
chr3_+_31574119
|
0.670
|
|
STT3B
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr14_-_95623665
|
0.667
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr19_-_51017124
|
0.662
|
NM_001114598
|
ASPDH
|
aspartate dehydrogenase domain containing
|
|
chr4_+_159593233
|
0.659
|
NM_004453
|
ETFDH
|
electron-transferring-flavoprotein dehydrogenase
|
|
chr4_-_7069801
|
0.637
|
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli)
|
|
chr12_-_51422012
|
0.636
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr1_+_45097669
|
0.620
|
|
RNF220
|
ring finger protein 220
|
|
chr5_+_1801493
|
0.601
|
NM_004553
|
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase)
|
|
chr8_-_124286566
|
0.598
|
NM_001204180
NM_001017926
NM_007222
|
ZHX1-C8ORF76
ZHX1
|
ZHX1-C8ORF76 readthrough
zinc fingers and homeoboxes 1
|
|
chr1_-_40042519
|
0.592
|
NM_001135653
NM_001135654
NM_003819
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr12_+_96252770
|
0.591
|
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr17_+_7123149
|
0.589
|
NM_000018
NM_001033859
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr12_+_67663364
|
0.583
|
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr7_-_65447233
|
0.581
|
NM_000181
|
GUSB
|
glucuronidase, beta
|
|
chr17_+_26800657
|
0.573
|
NM_001145975
NM_001145976
NM_003984
|
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr7_-_72936447
|
0.570
|
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr12_+_107168336
|
0.557
|
NM_018157
|
RIC8B
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr12_+_69202796
|
0.538
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr4_-_4543719
|
0.528
|
|
STX18
|
syntaxin 18
|
|
chr17_+_7531275
|
0.520
|
|
SHBG
|
sex hormone-binding globulin
|
|
chr16_-_4401265
|
0.516
|
NM_016069
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae)
|
|
chr2_+_28113612
|
0.510
|
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr2_-_198364522
|
0.508
|
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin)
|
|
chr12_+_14538232
|
0.506
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr11_+_86748870
|
0.504
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chrX_-_18372777
|
0.499
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr1_+_78354199
|
0.496
|
NM_001172309
NM_144573
|
NEXN
|
nexilin (F actin binding protein)
|
|
chr8_+_96037186
|
0.496
|
NM_152416
|
C8orf38
|
chromosome 8 open reading frame 38
|
|
chr19_+_50016606
|
0.495
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_-_72936592
|
0.488
|
NM_032408
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr7_-_65447206
|
0.481
|
|
GUSB
|
glucuronidase, beta
|
|
chr7_-_139763520
|
0.477
|
NM_022750
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr12_+_67663147
|
0.471
|
|
CAND1
|
cullin-associated and neddylation-dissociated 1
|
|
chr5_+_14664764
|
0.456
|
NM_138348
|
FAM105B
|
family with sequence similarity 105, member B
|
|
chr1_+_45274153
|
0.441
|
NM_001136537
|
BTBD19
|
BTB (POZ) domain containing 19
|
|
chr11_+_118230348
|
0.429
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr2_-_220034706
|
0.424
|
NM_001144889
NM_001144890
NM_144712
|
SLC23A3
|
solute carrier family 23 (nucleobase transporters), member 3
|
|
chr9_-_116138340
|
0.418
|
NM_031219
|
HDHD3
|
haloacid dehalogenase-like hydrolase domain containing 3
|
|
chr6_+_96025343
|
0.411
|
NM_024641
|
MANEA
|
mannosidase, endo-alpha
|
|
chr14_-_21566497
|
0.411
|
|
ZNF219
|
zinc finger protein 219
|
|
chr7_-_65447162
|
0.410
|
|
GUSB
|
glucuronidase, beta
|
|
chr7_-_25164902
|
0.399
|
NM_018947
|
CYCS
|
cytochrome c, somatic
|
|
chr19_+_44100734
|
0.399
|
NM_001145347
|
ZNF576
|
zinc finger protein 576
|
|
chr1_-_120484230
|
0.398
|
|
NOTCH2
|
notch 2
|
|
chr1_-_160313029
|
0.398
|
|
COPA
|
coatomer protein complex, subunit alpha
|
|
chr1_-_160312959
|
0.393
|
|
|
|
|
chr22_-_44893436
|
0.393
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr7_-_25164869
|
0.391
|
|
CYCS
|
cytochrome c, somatic
|
|
chr2_-_159312955
|
0.387
|
NM_001171637
NM_138803
|
CCDC148
|
coiled-coil domain containing 148
|
|
chr12_-_118810550
|
0.384
|
|
TAOK3
|
TAO kinase 3
|
|
chr11_+_118230320
|
0.381
|
|
UBE4A
|
ubiquitination factor E4A
|
|
chr20_-_36661822
|
0.367
|
NM_014657
|
TTI1
|
TELO2 interacting protein 1
|
|
chr4_-_83931861
|
0.362
|
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr15_-_82554990
|
0.359
|
|
EFTUD1
|
elongation factor Tu GTP binding domain containing 1
|
|
chr12_+_96252705
|
0.358
|
NM_003095
|
SNRPF
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr6_+_7108184
|
0.358
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chrX_-_64254592
|
0.355
|
NM_001178032
NM_001243804
|
ZC4H2
|
zinc finger, C4H2 domain containing
|
|
chr2_+_73441344
|
0.355
|
NM_006062
|
SMYD5
|
SMYD family member 5
|
|
chr20_+_44486219
|
0.355
|
NM_080752
|
ZSWIM3
|
zinc finger, SWIM-type containing 3
|
|
chr11_-_73720281
|
0.353
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr20_+_54967572
|
0.351
|
NM_001324
|
CSTF1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa
|
|
chr4_-_83931973
|
0.349
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr1_-_40042415
|
0.342
|
|
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form)
|
|
chr16_-_30546145
|
0.339
|
NM_023931
|
ZNF747
|
zinc finger protein 747
|
|
chr22_-_44893179
|
0.338
|
|
LDOC1L
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr2_+_198365012
|
0.337
|
|
HSPE1
|
heat shock 10kDa protein 1 (chaperonin 10)
|
|
chr22_+_38201113
|
0.330
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr11_+_117049387
|
0.328
|
|
SIDT2
|
SID1 transmembrane family, member 2
|
|
chr19_-_41812991
|
0.328
|
|
|
|
|
chr2_+_26568991
|
0.328
|
|
EPT1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific)
|
|
chr2_+_28113481
|
0.317
|
NM_199193
NM_199194
NM_004899
NM_199191
NM_199192
|
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator)
|
|
chr11_+_117049838
|
0.314
|
NM_001040455
|
SIDT2
|
SID1 transmembrane family, member 2
|