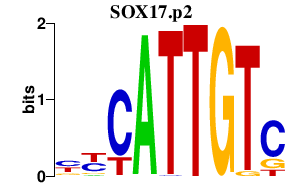
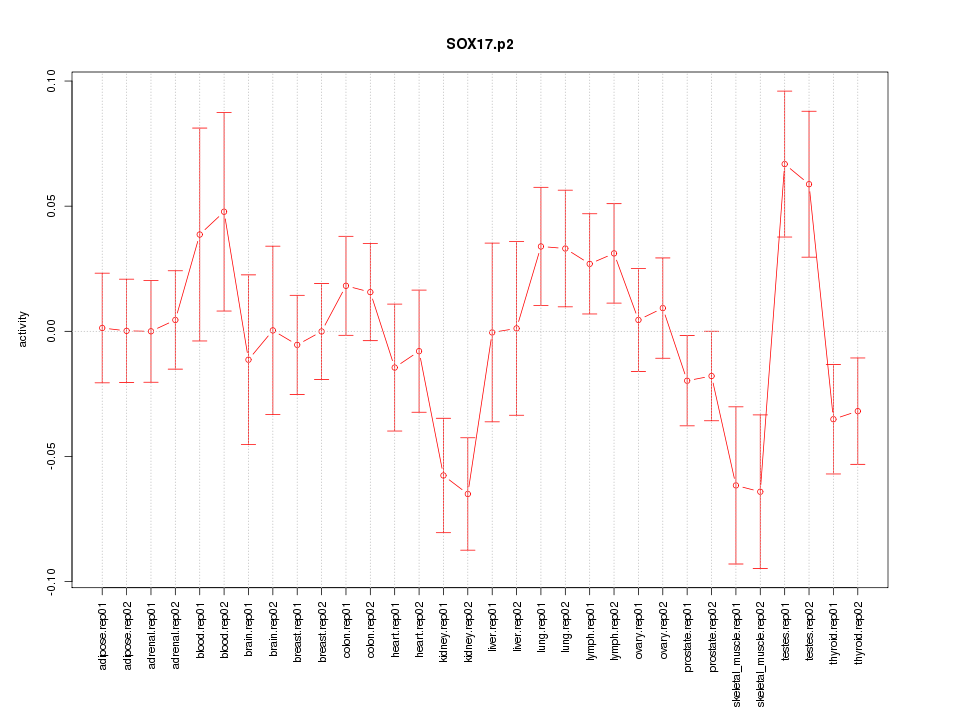
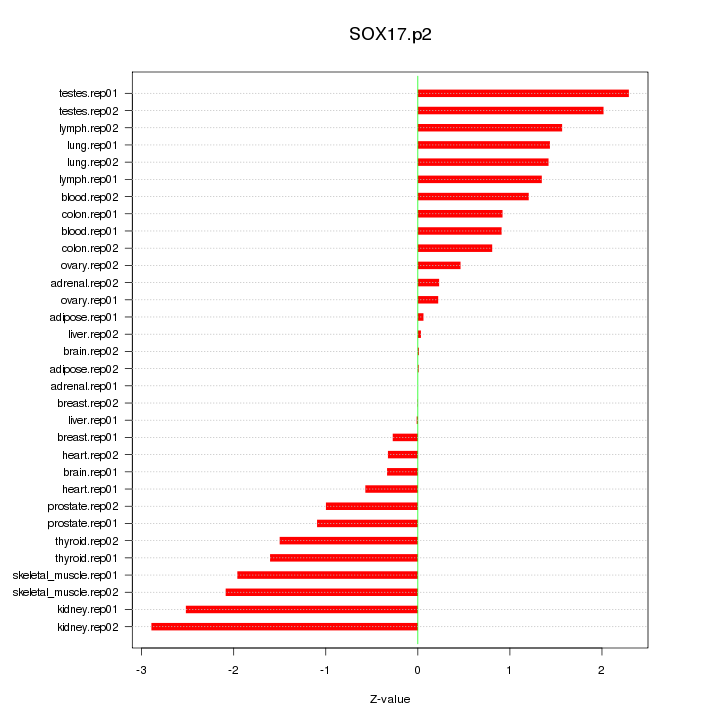
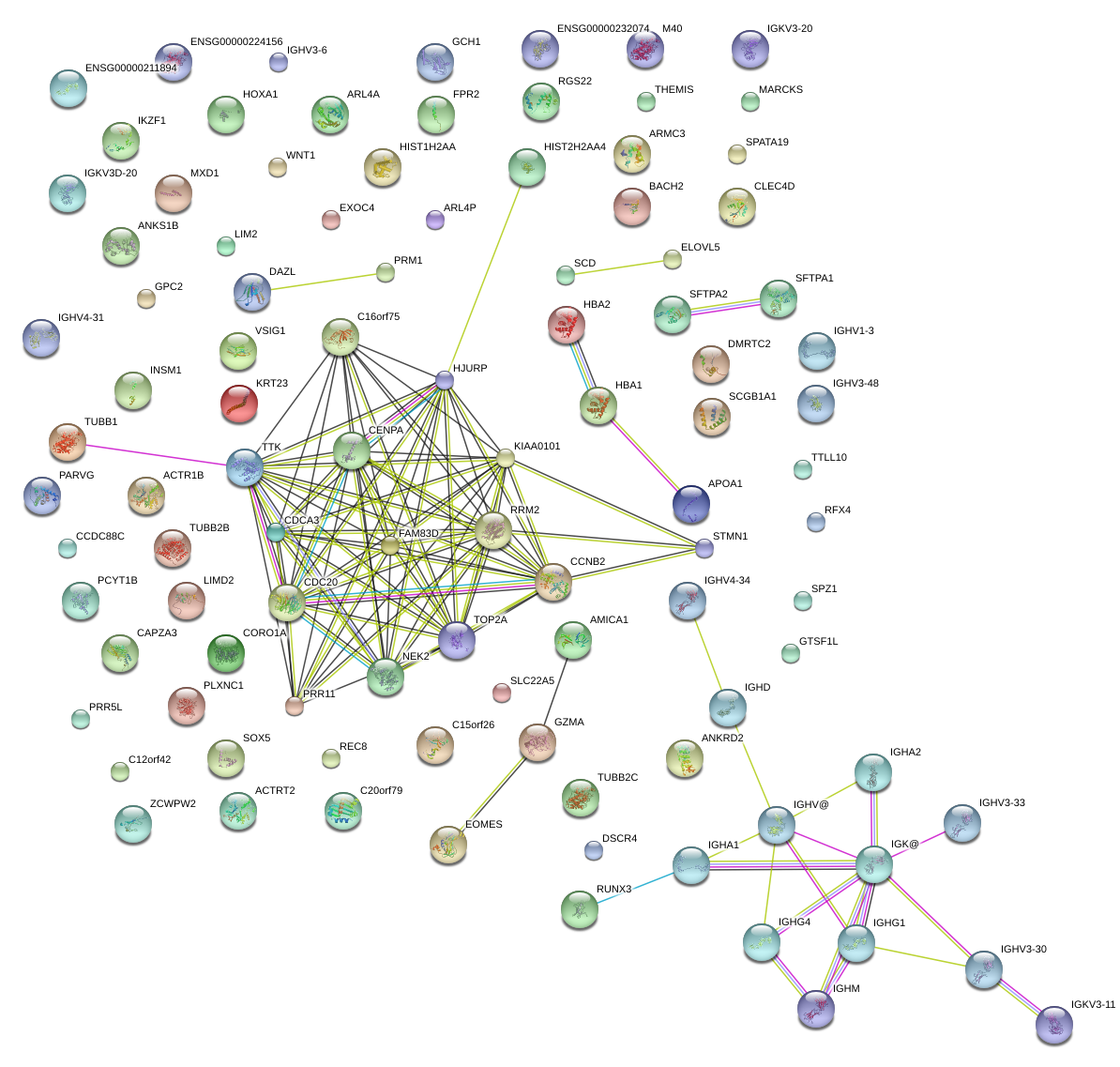
|
chr1_-_26232890
|
2.667
|
NM_001145454
NM_005563
|
STMN1
|
stathmin 1
|
|
chr14_-_106994001
|
2.471
|
|
IGHV3-48
IGH@
IGHA1
IGHM
|
immunoglobulin heavy variable 3-48
immunoglobulin heavy locus
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant mu
|
|
chr5_+_54398473
|
2.433
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chrX_-_24690740
|
2.399
|
NM_001163264
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr14_+_24641204
|
2.326
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr11_+_36317724
|
2.279
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr16_-_11375094
|
2.198
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr14_-_107218786
|
2.023
|
|
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr14_-_106725232
|
2.020
|
|
ZCWPW2
IGHV3-23
IGKV3-20
IGK@
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
immunoglobulin kappa locus
|
|
chr1_-_26232643
|
1.882
|
NM_203401
|
STMN1
|
stathmin 1
|
|
chr14_-_55369163
|
1.874
|
NM_000161
NM_001024024
NM_001024070
NM_001024071
|
GCH1
|
GTP cyclohydrolase 1
|
|
chr1_-_26233367
|
1.857
|
NM_203399
|
STMN1
|
stathmin 1
|
|
chr1_-_211848820
|
1.834
|
NM_001204182
NM_001204183
NM_002497
|
NEK2
|
NIMA (never in mitosis gene a)-related kinase 2
|
|
chr12_+_8666135
|
1.794
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr19_+_42349023
|
1.764
|
NM_001040283
|
DMRTC2
|
DMRT-like family C2
|
|
chr1_+_1115076
|
1.760
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr16_+_226678
|
1.759
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr20_+_18794369
|
1.752
|
NM_178483
|
C20orf79
|
chromosome 20 open reading frame 79
|
|
chr14_-_106926393
|
1.751
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr16_+_222845
|
1.726
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr17_-_61777478
|
1.711
|
NM_030576
|
LIMD2
|
LIM domain containing 2
|
|
chr20_+_37555055
|
1.642
|
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr14_-_106552313
|
1.636
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_23216953
|
1.605
|
NM_173081
|
ARMC3
|
armadillo repeat containing 3
|
|
chr6_-_128222175
|
1.587
|
NM_001010923
NM_001164685
|
THEMIS
|
thymocyte selection associated
|
|
chr7_-_99774821
|
1.563
|
NM_152742
|
GPC2
|
glypican 2
|
|
chr3_-_27764197
|
1.561
|
|
EOMES
|
eomesodermin
|
|
chr11_+_62186506
|
1.547
|
NM_003357
|
SCGB1A1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr20_-_42355566
|
1.544
|
NM_001008901
NM_176791
|
GTSF1L
|
gametocyte specific factor 1-like
|
|
chr15_+_59397276
|
1.529
|
NM_004701
|
CCNB2
|
cyclin B2
|
|
chr20_+_20348744
|
1.524
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr14_-_106725620
|
1.499
|
|
IGHV4-31
IGHV3-23
IGHA1
IGHG1
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 3-23
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_25291474
|
1.491
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_-_51891185
|
1.483
|
NM_001161748
NM_030657
|
LIM2
|
lens intrinsic membrane protein 2, 19kDa
|
|
chr1_+_43824598
|
1.472
|
NM_001255
|
CDC20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chr14_-_107131217
|
1.454
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr12_-_6960406
|
1.438
|
NM_031299
|
CDCA3
|
cell division cycle associated 3
|
|
chr14_-_91884138
|
1.434
|
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr11_-_118095708
|
1.431
|
NM_001098526
|
AMICA1
|
adhesion molecule, interacts with CXADR antigen 1
|
|
chr11_-_116708334
|
1.419
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr20_+_55099784
|
1.413
|
NM_001012971
|
FAM209A
FAM209B
|
family with sequence similarity 209, member A
family with sequence similarity 209, member B
|
|
chr14_+_24641084
|
1.401
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr12_-_103889745
|
1.398
|
NM_198521
NM_001099336
|
C12orf42
|
chromosome 12 open reading frame 42
|
|
chr2_+_70142152
|
1.397
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr6_-_53213699
|
1.385
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr6_-_25726754
|
1.382
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr14_-_91884164
|
1.377
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr20_+_37554954
|
1.371
|
NM_030919
|
FAM83D
|
family with sequence similarity 83, member D
|
|
chr17_-_61777467
|
1.362
|
|
LIMD2
|
LIM domain containing 2
|
|
chr7_+_50344255
|
1.351
|
NM_001220765
NM_001220766
NM_001220767
NM_001220768
NM_001220769
NM_001220770
NM_001220771
NM_001220772
NM_001220773
NM_001220774
NM_001220775
NM_001220776
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr17_-_39093671
|
1.339
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr19_+_52264452
|
1.339
|
NM_001005738
|
FPR2
|
formyl peptide receptor 2
|
|
chr12_-_24103953
|
1.336
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr17_-_39092714
|
1.309
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr12_+_18891040
|
1.302
|
NM_033328
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr16_+_30194925
|
1.294
|
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr22_+_44568835
|
1.281
|
NM_001137605
|
PARVG
|
parvin, gamma
|
|
chr17_+_57232859
|
1.270
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr17_-_38574042
|
1.264
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr16_+_11439279
|
1.240
|
NM_152308
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr7_-_27135524
|
1.239
|
NM_005522
NM_153620
|
HOXA1
|
homeobox A1
|
|
chr7_+_12727204
|
1.221
|
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr3_-_149510609
|
1.213
|
NM_001144960
|
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1
|
|
chr6_-_53213586
|
1.210
|
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr10_-_105992062
|
1.208
|
NM_025145
|
WDR96
|
WD repeat domain 96
|
|
chr6_-_15248886
|
1.202
|
|
|
|
|
chrX_+_107288199
|
1.202
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr2_+_10262853
|
1.201
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr5_+_79615789
|
1.188
|
NM_032567
|
SPZ1
|
spermatogenic leucine zipper 1
|
|
chr3_-_16646927
|
1.185
|
NM_001351
|
DAZL
|
deleted in azoospermia-like
|
|
chr6_+_80714321
|
1.184
|
NM_001166691
NM_003318
|
TTK
|
TTK protein kinase
|
|
chr20_+_57594308
|
1.182
|
NM_030773
|
TUBB1
|
tubulin, beta 1 class VI
|
|
chr2_+_70142236
|
1.179
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr12_+_49372235
|
1.153
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr12_-_99548471
|
1.144
|
NM_001204068
NM_001204069
NM_001204070
NM_020140
NM_181670
NM_001204066
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr8_-_101118170
|
1.143
|
|
RGS22
|
regulator of G-protein signaling 22
|
|
chr12_+_106976684
|
1.136
|
NM_213594
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr6_+_80714389
|
1.125
|
|
TTK
|
TTK protein kinase
|
|
chr6_-_91006460
|
1.125
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr1_+_2938045
|
1.122
|
NM_080431
|
ACTRT2
|
actin-related protein T2
|
|
chr11_-_133715368
|
1.121
|
NM_174927
|
SPATA19
|
spermatogenesis associated 19
|
|
chr15_+_81426605
|
1.121
|
NM_173528
|
C15orf26
|
chromosome 15 open reading frame 26
|
|
chr15_-_64673569
|
1.120
|
NM_001029989
NM_014736
|
KIAA0101
|
KIAA0101
|
|
chr2_+_27008881
|
1.117
|
NM_001809
NM_001042426
|
CENPA
|
centromere protein A
|
|
chr10_+_102106771
|
1.116
|
NM_005063
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase)
|
|
chr2_-_234763145
|
1.114
|
|
HJURP
|
Holliday junction recognition protein
|
|
chr10_+_81370694
|
1.098
|
NM_001093770
NM_001164644
NM_001164645
NM_001164646
NM_001164647
NM_005411
|
SFTPA1
|
surfactant protein A1
|
|
chr12_+_94542239
|
1.095
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr3_-_27763784
|
1.075
|
NM_005442
|
EOMES
|
eomesodermin
|
|
chr16_+_30194730
|
1.062
|
NM_001193333
NM_007074
|
CORO1A
|
coronin, actin binding protein, 1A
|
|
chr2_+_27008912
|
1.055
|
|
CENPA
|
centromere protein A
|
|
chr6_+_114178516
|
1.037
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr12_-_18890917
|
1.029
|
NM_033123
|
PLCZ1
|
phospholipase C, zeta 1
|
|
chr17_-_73150493
|
1.027
|
|
HN1
|
hematological and neurological expressed 1
|
|
chr8_-_60031545
|
1.026
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr2_-_2334887
|
1.022
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr14_-_106691698
|
1.020
|
|
|
|
|
chr2_+_10262694
|
1.011
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr14_-_106586333
|
1.010
|
|
IGHV4-31
|
immunoglobulin heavy variable 4-31
|
|
chr13_-_46756295
|
1.005
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_-_46756291
|
1.004
|
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr13_+_52436116
|
1.002
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chrX_-_145896248
|
1.000
|
NM_001144064
NM_001244892
|
CXorf51A
CXorf51B
|
chromosome X open reading frame 51A
chromosome X open reading frame 51B
|
|
chr12_-_11036815
|
0.999
|
NM_006250
|
PRH1
|
proline-rich protein HaeIII subfamily 1
|
|
chr11_-_125550674
|
0.998
|
NM_001612
NM_020069
NM_020107
NM_020108
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr12_+_11081833
|
0.998
|
NM_005042
NM_001110213
|
PRH2
|
proline-rich protein HaeIII subfamily 2
|
|
chr17_-_8113862
|
0.997
|
NM_004217
|
AURKB
|
aurora kinase B
|
|
chr1_+_1981902
|
0.994
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr11_+_560868
|
0.991
|
NM_001143993
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr21_-_32931289
|
0.986
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr13_+_46276445
|
0.985
|
NM_152719
|
SPERT
|
spermatid associated
|
|
chr20_+_44441247
|
0.984
|
NM_007019
NM_181799
NM_181800
NM_181802
NM_181803
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_-_175869910
|
0.982
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr22_-_37608352
|
0.980
|
NM_001051
|
SSTR3
|
somatostatin receptor 3
|
|
chr17_-_56565700
|
0.980
|
NM_001080439
|
HSF5
|
heat shock transcription factor family member 5
|
|
chr20_+_44441334
|
0.970
|
|
UBE2C
|
ubiquitin-conjugating enzyme E2C
|
|
chr8_-_67525426
|
0.969
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr20_+_55108265
|
0.965
|
NM_001013646
|
FAM209B
|
family with sequence similarity 209, member B
|
|
chr2_+_70142303
|
0.955
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr14_-_106573348
|
0.953
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr1_-_197115566
|
0.953
|
NM_001206846
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr7_-_100061893
|
0.953
|
NM_001004323
|
C7orf61
|
chromosome 7 open reading frame 61
|
|
chr2_-_89619820
|
0.949
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr12_-_24715349
|
0.943
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_560706
|
0.938
|
NM_173573
|
C11orf35
|
chromosome 11 open reading frame 35
|
|
chr2_+_75061234
|
0.930
|
|
HK2
|
hexokinase 2
|
|
chr17_-_78450247
|
0.929
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr16_+_11439311
|
0.925
|
|
RMI2
|
RMI2, RecQ mediated genome instability 2, homolog (S. cerevisiae)
|
|
chr12_-_11463353
|
0.909
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr7_+_18535351
|
0.907
|
NM_001204145
NM_001204146
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr12_-_68726036
|
0.906
|
NM_001205028
NM_001205029
NM_017440
NM_020128
|
MDM1
|
Mdm1 nuclear protein homolog (mouse)
|
|
chr7_+_120969089
|
0.904
|
NM_057168
|
WNT16
|
wingless-type MMTV integration site family, member 16
|
|
chr12_-_96390002
|
0.898
|
NM_002108
|
HAL
|
histidine ammonia-lyase
|
|
chr7_+_37779995
|
0.896
|
NM_181791
|
GPR141
|
G protein-coupled receptor 141
|
|
chr16_-_84651639
|
0.895
|
NM_021149
|
COTL1
|
coactosin-like 1 (Dictyostelium)
|
|
chr11_-_94964209
|
0.894
|
NM_144665
|
SESN3
|
sestrin 3
|
|
chr19_-_55677919
|
0.892
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr4_-_122744782
|
0.892
|
|
CCNA2
|
cyclin A2
|
|
chr3_-_20227618
|
0.886
|
NM_001012409
NM_001012410
NM_001012411
NM_001012412
NM_001012413
NM_138484
NM_001199251
NM_001199252
NM_001199253
NM_001199254
NM_001199255
NM_001199256
NM_001199257
|
SGOL1
|
shugoshin-like 1 (S. pombe)
|
|
chr6_-_53213747
|
0.878
|
NM_001242828
NM_001242830
NM_001242831
NM_021814
|
ELOVL5
|
ELOVL fatty acid elongase 5
|
|
chr9_+_135854097
|
0.874
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr19_+_58790306
|
0.863
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr7_+_27135983
|
0.859
|
|
|
|
|
chr13_+_53029614
|
0.858
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr21_+_34442446
|
0.856
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr14_-_106780535
|
0.856
|
|
IGHA1
IGHA2
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr14_-_107099379
|
0.856
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr3_-_160167600
|
0.853
|
NM_173084
|
TRIM59
|
tripartite motif containing 59
|
|
chr5_-_157079371
|
0.852
|
NM_007017
NM_178424
|
SOX30
|
SRY (sex determining region Y)-box 30
|
|
chr13_-_46756326
|
0.851
|
NM_002298
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin)
|
|
chr1_+_50571963
|
0.848
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_+_169923531
|
0.845
|
NM_199204
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr12_-_6961044
|
0.841
|
|
CDCA3
|
cell division cycle associated 3
|
|
chr2_+_232573249
|
0.839
|
|
PTMA
|
prothymosin, alpha
|
|
chr4_-_74864296
|
0.839
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr12_-_45270071
|
0.838
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr21_-_15918635
|
0.837
|
NM_022136
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr7_-_150675236
|
0.835
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr4_-_80993713
|
0.829
|
|
ANTXR2
|
anthrax toxin receptor 2
|
|
chr3_+_51422716
|
0.824
|
|
MANF
|
mesencephalic astrocyte-derived neurotrophic factor
|
|
chr5_+_148737569
|
0.824
|
NM_024028
|
PCYOX1L
|
prenylcysteine oxidase 1 like
|
|
chr5_+_176875325
|
0.822
|
NM_001174102
|
PRR7
|
proline rich 7 (synaptic)
|
|
chr10_+_12391728
|
0.822
|
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr19_-_6110561
|
0.817
|
NM_000635
NM_134433
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression)
|
|
chr2_+_149633173
|
0.815
|
|
|
|
|
chr15_+_76629101
|
0.814
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr8_-_145743151
|
0.808
|
NM_004260
|
RECQL4
|
RecQ protein-like 4
|
|
chr5_-_41071404
|
0.806
|
NM_173489
|
HEATR7B2
|
HEAT repeat family member 7B2
|
|
chr17_-_77478562
|
0.805
|
NM_001082575
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr18_+_21718930
|
0.804
|
NM_012189
NM_138643
NM_153768
NM_153769
NM_153770
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated
|
|
chr19_+_43919034
|
0.797
|
NM_001130011
|
TEX101
|
testis expressed 101
|
|
chr2_-_70995113
|
0.786
|
NM_001185055
NM_001617
NM_017482
NM_017488
|
ADD2
|
adducin 2 (beta)
|
|
chr20_+_9495297
|
0.783
|
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr16_+_19467771
|
0.782
|
NM_024780
|
TMC5
|
transmembrane channel-like 5
|
|
chr3_+_122334461
|
0.773
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr15_+_74833517
|
0.770
|
NM_006465
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like)
|
|
chr9_+_82187599
|
0.763
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr10_+_99349354
|
0.762
|
NM_001009997
|
C10orf62
|
chromosome 10 open reading frame 62
|
|
chr1_+_158259558
|
0.762
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr20_+_9494995
|
0.760
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr10_-_127371593
|
0.759
|
NM_001128202
|
C10orf122
|
chromosome 10 open reading frame 122
|
|
chr8_-_82024277
|
0.758
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr7_+_128349111
|
0.757
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr16_+_4784446
|
0.757
|
|
C16orf71
|
chromosome 16 open reading frame 71
|
|
chr7_+_69064580
|
0.757
|
|
AUTS2
|
autism susceptibility candidate 2
|
|
chr6_-_30655078
|
0.754
|
NM_001134870
|
PPP1R18
|
protein phosphatase 1, regulatory subunit 18
|
|
chr16_+_15744082
|
0.754
|
NM_017668
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr11_+_196760
|
0.746
|
NM_053280
|
ODF3
|
outer dense fiber of sperm tails 3
|
|
chr2_-_234763182
|
0.744
|
NM_018410
|
HJURP
|
Holliday junction recognition protein
|
|
chr20_+_207898
|
0.743
|
NM_080831
|
DEFB129
|
defensin, beta 129
|
|
chr16_+_2016874
|
0.741
|
NM_174903
|
RNF151
|
ring finger protein 151
|
|
chr22_-_51001327
|
0.740
|
NM_001123225
|
SYCE3
|
synaptonemal complex central element protein 3
|
|
chr9_+_82187469
|
0.737
|
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr1_+_34632483
|
0.734
|
NM_032884
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr22_-_24181173
|
0.732
|
NM_001002862
NM_001135751
NM_198440
|
DERL3
|
Der1-like domain family, member 3
|
|
chrX_+_145891301
|
0.726
|
NM_001144064
NM_001244892
|
CXorf51A
CXorf51B
|
chromosome X open reading frame 51A
chromosome X open reading frame 51B
|
|
chr2_+_232573226
|
0.724
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr15_+_40886442
|
0.723
|
NM_144508
NM_170589
|
CASC5
|
cancer susceptibility candidate 5
|
|
chr7_-_122339186
|
0.721
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr11_+_63580922
|
0.717
|
NM_138471
|
C11orf84
|
chromosome 11 open reading frame 84
|